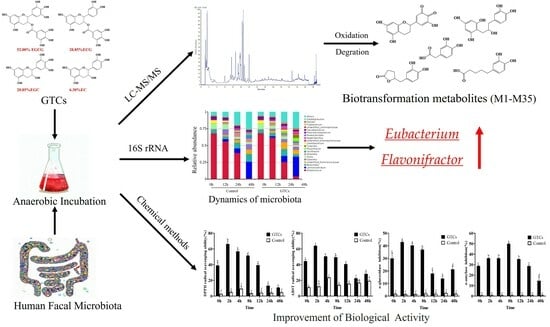
Microbial-Transferred Metabolites and Improvement of Biological Activities of Green Tea Catechins by Human Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Sample Preparation
2.3. Fecal Sample Collection and Biotransformation of GTCs by Human Gut Microbiota
2.4. Analysis of the Microbial Metabolites of GTCs via UHPLC-Q-Orbitrap-MS/MS
2.5. Analytical Strategy Based on UHPLC-Orbitrap MS/MS
2.6. Antioxidant Assay
2.7. Measurement of α-Amylase and α-Glucosidase Inhibitory Activity
2.8. Analysis of Gut Microbiota Composition
2.9. Data Analysis
3. Results
3.1. Identification of the Metabolites of GTCs Fermented via Human Fecal Fermentation In Vitro
3.2. Microbial Biotransformation Pathways of GTCs during In Vitro Human Fecal Fermentation
3.3. Dynamic Enhancements in Antioxidant, α-Glucosidase and α-Amylase Inhibitory Activity of GTCs during Fermentation
3.4. Dynamics of Microbiota during the Fermentation Process
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Suzuki, Y.; Miyoshi, N.; Isemura, M. Health-Promoting Effects of Green Tea. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2012, 88, 88–101. [Google Scholar] [CrossRef]
- Annunziata, G.; Maisto, M.; Schisano, C.; Ciampaglia, R.; Daliu, P.; Narciso, V.; Tenore, G.; Novellino, E. Colon Bioaccessibility and Antioxidant Activity of White, Green and Black Tea Polyphenols Extract after In Vitro Simulated Gastrointestinal Digestion. Nutrients 2018, 10, 1711. [Google Scholar] [CrossRef] [PubMed]
- Butt, M.S.; Ahmad, R.S.; Sultan, M.T.; Qayyum, M.M.N.; Naz, A. Green tea and anticancer perspectives: Updates from last decade. Crit. Rev. Food Sci. Nutr. 2015, 55, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.P.; Sugita, M.; Fukuzawa, Y.; Timm, D.; Ozeki, M.; Okubo, T. Green Tea Catechin Association with Ultraviolet Radiation-Induced Erythema: A Systematic Review and Meta-Analysis. Molecules 2021, 26, 3702. [Google Scholar] [CrossRef] [PubMed]
- Castaldo, L.; Toriello, M.; Sessa, R.; Izzo, L.; Lombardi, S.; Narváez, A.; Ritieni, A.; Grosso, M. Antioxidant and Anti-Inflammatory Activity of Coffee Brew Evaluated after Simulated Gastrointestinal Digestion. Nutrients 2021, 13, 4368. [Google Scholar] [CrossRef]
- Higdon, J.V.; Frei, B. Tea catechins and polyphenols: Health effects, metabolism, and antioxidant functions. Crit. Rev. Food Sci. Nutr. 2003, 43, 89–143. [Google Scholar] [CrossRef]
- Musial, C.; Kuban-Jankowska, A.; Gorska-Ponikowska, M. Beneficial Properties of Green Tea Catechins. Int. J. Mol. Sci. 2020, 21, 1744. [Google Scholar] [CrossRef]
- Kochman, J.; Jakubczyk, K.; Antoniewicz, J.; Mruk, H.; Janda, K. Health Benefits and Chemical Composition of Matcha Green Tea: A Review. Molecules 2020, 26, 85. [Google Scholar] [CrossRef]
- Borges, G.; van der Hooft, J.J.J.; Crozier, A. A comprehensive evaluation of the [2-14C](-)-epicatechin metabolome in rats. Free Radic. Biol. Med. 2016, 99, 128–138. [Google Scholar] [CrossRef]
- Mena, P.; Bresciani, L.; Brindani, N.; Ludwig, I.A.; Pereira-Caro, G.; Angelino, D.; Llorach, R.; Calani, L.; Brighenti, F.; Clifford, M.N.; et al. Phenyl-γ-valerolactones and phenylvaleric acids, the main colonic metabolites of flavan-3-ols: Synthesis, analysis, bioavailability, and bioactivity. Nat. Prod. Rep. 2019, 36, 714–752. [Google Scholar] [CrossRef]
- Li, Q.; Van Herreweghen, F.; Onyango, S.O.; De Mey, M.; Van de Wiele, T. In Vitro Microbial Metabolism of (+)-Catechin Reveals Fast and Slow Converters with Individual-Specific Microbial and Metabolite Markers. J. Agric. Food Chem. 2022, 70, 10405–10416. [Google Scholar] [CrossRef] [PubMed]
- Al-Ishaq, R.K.; Liskova, A.; Kubatka, P.; Büsselberg, D. Enzymatic Metabolism of Flavonoids by Gut Microbiota and Its Impact on Gastrointestinal Cancer. Cancers 2021, 13, 3934. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Bao, T.; Zhang, M.; Zhou, Y.; Chen, W. Phenolic profile of jujube fruit subjected to gut microbiota fermentation and its antioxidant potential against ethyl carbamate-induced oxidative damage. J. Zhejiang Univ. Sci. B 2021, 22, 397–409. [Google Scholar] [CrossRef]
- Yan, Y.; Fu, C.; Cui, X.; Pei, X.; Li, A.; Qin, X.; Du, C.; Du, H. Metabolic profile and underlying antioxidant improvement of Ziziphi Spinosae Folium by human intestinal bacteria. Food Chem. 2020, 320, 126651. [Google Scholar] [CrossRef]
- Kc, D.; Sumner, R.; Lippmann, S. Gut microbiota and health. Postgrad. Med. 2020, 132, 274. [Google Scholar] [CrossRef]
- Zafar, H.; Saier, M.H. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef]
- Li, Y.; Rahman, S.U.; Huang, Y.; Zhang, Y.; Ming, P.; Zhu, L.; Chu, X.; Li, J.; Feng, S.; Wang, X.; et al. Green tea polyphenols decrease weight gain, ameliorate alteration of gut microbiota, and mitigate intestinal inflammation in canines with high-fat-diet-induced obesity. J. Nutr. Biochem. 2020, 78, 108324. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Su, Y.; Hu, K.; Li, D.; Guo, H.; Xie, Z. Microbial-Transferred Metabolites of Black Tea Theaflavins by Human Gut Microbiota and Their Impact on Antioxidant Capacity. Molecules 2023, 28, 5871. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; de Bruijn, W.J.C.; Bruins, M.E.; Vincken, J.-P. Reciprocal Interactions between Epigallocatechin-3-gallate (EGCG) and Human Gut Microbiota In Vitro. J. Agric. Food Chem. 2020, 68, 9804–9815. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Ma, Y.; Liang, C.; Gao, J.; Wang, H.; Zhang, L. A Systematic Study of the Metabolites of Dietary Acacetin in Vivo and in Vitro Based on UHPLC-Q-TOF-MS/MS Analysis. J. Agric. Food Chem. 2019, 67, 5530–5543. [Google Scholar] [CrossRef]
- Wang, B.; Lu, Y.; Hu, X.; Feng, J.; Shen, W.; Wang, R.; Wang, H. Systematic Strategy for Metabolites of Amentoflavone In Vivo and In Vitro Based on UHPLC-Q-TOF-MS/MS Analysis. J. Agric. Food Chem. 2020, 68, 14808–14823. [Google Scholar] [CrossRef]
- Li, M.; Luo, X.; Ho, C.-T.; Li, D.; Guo, H.; Xie, Z. A new strategy for grading of Lu’an guapian green tea by combination of differentiated metabolites and hypoglycaemia effect. Food Res. Int. 2022, 159, 111639. [Google Scholar] [CrossRef]
- Zahid, H.F.; Ali, A.; Ranadheera, C.S.; Fang, Z.; Ajlouni, S. Identification of Phenolics Profile in Freeze-Dried Apple Peel and Their Bioactivities during In Vitro Digestion and Colonic Fermentation. Int. J. Mol. Sci. 2023, 24, 1514. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mao, B.; Cui, S.; Zhang, Q.; Zhao, J.; Tang, X.; Chen, W. Absorption, metabolism, bioactivity, and biotransformation of epigallocatechin gallate. Crit. Rev. Food Sci. Nutr. 2023, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Favari, C.; Mena, P.; Curti, C.; Rio, D.; Angelino, D. Flavan-3-ols: Catechins and Proanthocyanidins. In Dietary Polyphenols; Tomás-Barberán, F.A., González-Sarrías, A., García-Villalba, R., Eds.; Wiley: Hoboken, NJ, USA, 2020; pp. 283–317. ISBN 978-1-119-56375-4. [Google Scholar]
- Han, X.-D.; Zhang, Y.-Y.; Wang, K.-L.; Huang, Y.-P.; Yang, Z.-B.; Liu, Z. The involvement of Nrf2 in the protective effects of (-)-Epigallocatechin-3-gallate (EGCG) on NaAsO2-induced hepatotoxicity. Oncotarget 2017, 8, 65302–65312. [Google Scholar] [CrossRef] [PubMed]
- Absorption, Metabolism, Anti-Cancer Effect and Molecular Targets of Epigallocatechin Gallate (EGCG): An Updated Review-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/27645804/ (accessed on 12 August 2023).
- Cao, H.; Chen, X.; Jassbi, A.R.; Xiao, J. Microbial biotransformation of bioactive flavonoids. Biotechnol. Adv. 2015, 33, 214–223. [Google Scholar] [CrossRef]
- Wang, L.Q.; Meselhy, M.R.; Li, Y.; Nakamura, N.; Min, B.S.; Qin, G.W.; Hattori, M. The heterocyclic ring fission and dehydroxylation of catechins and related compounds by Eubacterium sp. strain SDG-2, a human intestinal bacterium. Chem. Pharm. Bull. 2001, 49, 1640–1643. [Google Scholar] [CrossRef] [PubMed]
- Meselhy, M.R.; Nakamura, N.; Hattori, M. Biotransformation of (-)-epicatechin 3-O-gallate by human intestinal bacteria. Chem. Pharm. Bull. 1997, 45, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Takagaki, A.; Nanjo, F. Metabolism of (-)-epigallocatechin gallate by rat intestinal flora. J. Agric. Food Chem. 2010, 58, 1313–1321. [Google Scholar] [CrossRef]
- Stoupi, S.; Williamson, G.; Drynan, J.W.; Barron, D.; Clifford, M.N. A comparison of the in vitro biotransformation of (-)-epicatechin and procyanidin B2 by human faecal microbiota. Mol. Nutr. Food Res. 2010, 54, 747–759. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, J.; Duan, H.; Li, R.; Peng, W.; Wu, C. Activation of Nrf2/HO-1 signaling: An important molecular mechanism of herbal medicine in the treatment of atherosclerosis via the protection of vascular endothelial cells from oxidative stress. J. Adv. Res. 2021, 34, 43–63. [Google Scholar] [CrossRef]
- Chen, W.; Zhu, X.; Lu, Q.; Zhang, L.; Wang, X.; Liu, R. C-ring cleavage metabolites of catechin and epicatechin enhanced antioxidant activities through intestinal microbiota. Food Res. Int. 2020, 135, 109271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-Q.; Lin, L.-L.; Xu, H.-J. Research on antioxidant performance of diglucosyl gallic acid and its application in emulsion cosmetics. Int. J. Cosmet. Sci. 2022, 44, 177–188. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Lv, Y.; Yao, K. Effects of tea polyphenols on the activities of α-amylase, pepsin, trypsin and lipase. Food Chem. 2007, 101, 1178–1182. [Google Scholar] [CrossRef]
- Zhu, S.; Li, J.; Li, W.; Li, S.; Yang, X.; Liu, X.; Sun, L. Enzymic catalyzing affinity to substrate affects inhibitor-enzyme binding interactions: Inhibition behaviors of EGCG against starch digestion by individual and co-existing α-amylase and amyloglucosidase. Food Chem. 2022, 388, 133047. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fowler, M.I.; Messenger, D.J.; Ordaz-Ortiz, J.J.; Gu, X.; Shi, S.; Terry, L.A.; Berry, M.J.; Lian, G.; Wang, S. Inhibition of the intestinal postprandial glucose transport by gallic acid and gallic acid derivatives. Food Funct. 2021, 12, 5399–5406. [Google Scholar] [CrossRef] [PubMed]
- Selma, M.V.; Espín, J.C.; Tomás-Barberán, F.A. Interaction between phenolics and gut microbiota: Role in human health. J. Agric. Food Chem. 2009, 57, 6485–6501. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Ho, C.-T.; Zhang, X. Interaction between Tea Polyphenols and Intestinal Microbiota in Host Metabolic Diseases from the Perspective of the Gut-Brain Axis. Mol. Nutr. Food Res. 2020, 64, e2000187. [Google Scholar] [CrossRef]
- Li, J.; Chen, C.; Yang, H.; Yang, X. Tea polyphenols regulate gut microbiota dysbiosis induced by antibiotic in mice. Food Res. Int. 2021, 141, 110153. [Google Scholar] [CrossRef]
- Folz, J.; Culver, R.N.; Morales, J.M.; Grembi, J.; Triadafilopoulos, G.; Relman, D.A.; Huang, K.C.; Shalon, D.; Fiehn, O. Human metabolome variation along the upper intestinal tract. Nat. Metab. 2023, 5, 777–788. [Google Scholar] [CrossRef] [PubMed]




| No. | RT. | Tentative Identification | Formula | Theoretical Mass m/z | Experimental Mass m/z | Error (ppm) | MS/MS Fragments |
|---|---|---|---|---|---|---|---|
| M0 | 8.62 | EGCG a | C22H18O11 | 458.08545 | 458.08546 | 0.02 | 125.02437, 169.01418 |
| M1 | 7.64 | EGC a | C15H14O7 | 306.07450 | 306.07447 | −0.10 | 125.02433, 137.02422, 167.03477, 121.02950, 109.02927 |
| M2 | 9.74 | ECG a | C22H18O10 | 442.09054 | 442.09046 | −0.18 | 125.02451, 169.01428, 109.0259, 231.06375 |
| M3 | 8.66 | EC a | C15H14O6 | 290.07958 | 290.07963 | 0.17 | 109.02954, 123.04526, 245.08153, 151.04005 |
| M4 | 3.45 | gallic acid a | C7H6O5 | 170.02207 | 170.02204 | −0.18 | 169.01439, 125.02456, 69.03470, 97.02968, 79.01898 |
| M5 | 3.67 | Pyrogallol a | C6H6O3 | 126.03224 | 126.03230 | 0.48 | 125.02448, 97.02959, 81.0364, 79.01917, 69.03465 |
| M6 | 7.64 | EGCG quinone | C22H16O11 | 456.06926 | 456.06992 | 1.45 | 125.02444, 169.01413, 137.02463, 149.01947, 285.04010, 161.02415 |
| M7 | 9.75 | EGC quinone | C15H12O7 | 304.05830 | 304.05894 | 2.10 | 137.02446, 125.02437, 165.01947, 137.02453, 285.04010, 303.0507 |
| M8 | 8.61 | ECG quinone | C22H18O10 | 440.07435 | 440.07484 | 1.11 | 125.02437, 137.02455, 161.02403, 169.01920, 149.02425 |
| M9 | 8.67 | EC quinone | C15H12O6 | 288.06338 | 288.06397 | 2.05 | 125.02450, 161.02403, 137.02452, 149.02425, 269.04572 |
| M10 | 14.01 | EGCG+O | C22H18O12 | 474.07983 | 474.07951 | −0.67 | 125.02402, 165.01817, 169.01385 |
| M11 | 8.73 | 1-(3′,4′-Dihydroxyphenyl)-3-(2″,4″,6″-Trihydroxyphenyl)-propan-2-ol | C15H16O6 | 292.09523 | 292.09528 | 0.17 | 123.04530, 135.04524, 167.03503, 247.09755 |
| M12 | 8.40 | 1-(3′,5′-Dihydroxyphenyl)-3-(2″,4″,6″-Trihydroxyphenyl)-propan-2-ol | C15H16O6 | 292.09523 | 292.09534 | 0.38 | 123.04530, 167.03503, 247.09755, 205.08794, 139.0837, 109.02961 |
| M13 | 9.97 | 1-(3′-droxyphenyl)-3-(2″,4″,6″-Trihydroxyphenyl)-propan-2-ol | C15H16O5 | 276.09970 | 276.10032 | −0.22 | 107.05042, 231.10236, 167.07207, 189.09196, 147.08142 |
| M14 | 7.82 | 1-(3′,4′,5′-Trihydroxyphenyl)-3-(2″,4″,6″-Trihydroxyphenyl)-propan-2-ol | C15H16O7 | 308.09015 | 308.09010 | −0.17 | 139.04012, 263.22664, 167.03503, 125.02448, 221.08154 |
| M15 | 7.22 | 1-(3′,4′-Dihydroxyphenyl)-3- (2″,4″,6″-Trihydroxyphenyl)-propan-2-yl gallate | C22H20O10 | 444.10564 | 444.10410 | −3.47 | 125.02464, 169.01437, 291.07657, 245.08110, 137.02432 |
| M16 | 14.01 | ECG –O3 | C22H20O7 | 396.12090 | 396.12049 | −1.04 | 125.02454, 167.03506, 121.02946, 205.04964, 139.02444, 109.02969 |
| M17 | 9.15 | 5-(2′-Hydroxyphenyl)-γ-valerolactone | C11H12O3 | 192.07919 | 192.07922 | 0.26 | 147.08154, 106.04253, 121.02972, 102.02972, 107.04990 |
| M18 | 9.83 | 5-(3′-Hydroxyphenyl)-γ-valerolactone or its isomers | C11H12O3 | 192.07919 | 192.07929 | 0.51 | 147.08163, 105.93622, 123.02972, 102.94908, 107.05032 |
| M19 | 10.81 | 5-(3′-Hydroxyphenyl)-γ-valerolactones or its isomers | C11H12O3 | 192.07919 | 192.07908 | −0.59 | 147.08177, 106.04253, 121.02988, 102.94888, 191.07159 |
| M20 | 9.28 | 5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone | C11H12O4 | 208.07410 | 208.07407 | −0.14 | 123.04532, 163.07660, 122.03742, 207.06624, 81.03461 |
| M21 | 8.68 | 5-(3′,5′-Dihydroxyphenyl)-γ-valerolactone | C11H12O4 | 208.07410 | 208.07403 | −0.34 | 123.04528, 163.07646, 122.03754, 81.03459, 79.05441 |
| M22 | 7.85 | 5-(3′,4′,5′-Trihydroxyphenyl)-γ-valerolactone | C11H12O5 | 224.06902 | 224.06902 | 0.00 | 243.06890, 179.07138, 123.04522, 133.06598, 122.03753, 161.06102 |
| M23 | 12.01 | 5-(2′-Hydroxyphenyl) -γ-valeric acid or its isomers | C11H14O3 | 194.09484 | 194.09475 | −0.46 | 193.08685, 175.07658, 149.09723, 106.04269, 121.02948 |
| M24 | 9.15 | 5-(3′,5′-Dihydroxyphenyl)-γ-valeric acid | C11H14O4 | 210.08975 | 210.08964 | −0.52 | 191.07146, 165.09203, 101.02448, 107.05035, 147.08165 |
| M25 | 9.81 | 5-(3′,4′-Dihydroxyphenyl)-γ-valeric acid | C11H14O4 | 210.08975 | 210.08964 | −0.52 | 123.08171, 81.032487, 107.05025, 149.06085, 147.08154, 91.05512 |
| M26 | 7.91 | 5-(3′,4′,5′-Trihydroxyphenyl)-γ-valeric acid | C11H14O5 | 226.08467 | 226.08460 | −0.31 | 179.07138, 123.04532, 81.03457, 101.02434 |
| M27 | 7.60 | 4-Hydroxyphenylbutyric acid | C10H12O3 | 180.07864 | 180.07918 | 2.94 | 179.07130, 134.98804, 90.99820, 04.92816 |
| M28 | 9.28 | 4-phenylbutyric acid a | C10H12O2 | 164.08427 | 164.08421 | −0.37 | 163.07648, 121.06597, 81.03464, 145.89063 |
| M29 | 11.97 | 3-phenylpropionic acid a | C9H10O2 | 150.06862 | 150.06852 | −0.67 | 149.06081, 105.07104, 123.46254, 103.05509 |
| M30 | 8.81 | 3-(3′,4′-Dihydroxyphenyl)propanoic acid a | C9H10O4 | 182.05845 | 182.05851 | 0.33 | 181.05078, 112.98579, 92.99387, 136.98322 |
| M31 | 11.12 | phenylacetic acid a | C8H8O2 | 136.05297 | 136.05306 | 0.66 | 135.04448, 91.05541, 67.72299 |
| M32 | 8.50 | 2-(4′-Hydroxyphenyl)acetic acid a | C8H8O3 | 152.04789 | 152.04787 | −0.13 | 107.05036 |
| M33 | 9.84 | 2-(3′-Hydroxyphenyl)acetic acid | C8H8O3 | 152.04789 | 152.04777 | −0.79 | 151.04015, 107.05034 |
| M34 | 7.77 | 2-(3′,4′-Dihydroxyphenyl)acetic acid a | C8H8O4 | 168.04280 | 168.04267 | −0.77 | 123.04521, 95.05029 |
| M35 | 7.84 | 4-Hydroxybenzoic acid a | C7H6O3 | 138.03224 | 138.03216 | −0.58 | 137.06108, 93.03466, 85.05035 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Y.; Hu, K.; Li, D.; Guo, H.; Sun, L.; Xie, Z. Microbial-Transferred Metabolites and Improvement of Biological Activities of Green Tea Catechins by Human Gut Microbiota. Foods 2024, 13, 792. https://doi.org/10.3390/foods13050792
Su Y, Hu K, Li D, Guo H, Sun L, Xie Z. Microbial-Transferred Metabolites and Improvement of Biological Activities of Green Tea Catechins by Human Gut Microbiota. Foods. 2024; 13(5):792. https://doi.org/10.3390/foods13050792
Chicago/Turabian StyleSu, You, Kaiyin Hu, Daxiang Li, Huimin Guo, Li Sun, and Zhongwen Xie. 2024. "Microbial-Transferred Metabolites and Improvement of Biological Activities of Green Tea Catechins by Human Gut Microbiota" Foods 13, no. 5: 792. https://doi.org/10.3390/foods13050792
APA StyleSu, Y., Hu, K., Li, D., Guo, H., Sun, L., & Xie, Z. (2024). Microbial-Transferred Metabolites and Improvement of Biological Activities of Green Tea Catechins by Human Gut Microbiota. Foods, 13(5), 792. https://doi.org/10.3390/foods13050792