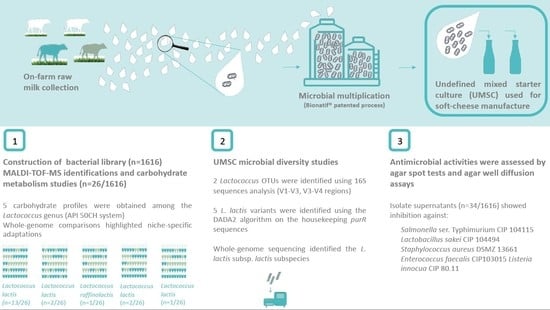
Lactococcus lactis Diversity Revealed by Targeted Amplicon Sequencing of purR Gene, Metabolic Comparisons and Antimicrobial Properties in an Undefined Mixed Starter Culture Used for Soft-Cheese Manufacture
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Gene Sequencing of the UMSC Microbiota
2.2.1. DNA Extraction
2.2.2. Gene Sequencing and Data Analysis of 16S rRNA
2.2.3. purR Gene Sequencing and Data Analysis
2.3. Bacterial Library
2.4. Phenotypic Profiles and LAB Isolates Identification
2.4.1. Bacterial Carbohydrate Metabolism Profile
2.4.2. MALDI-TOF MS Bacterial Identification
2.4.3. DNA Extraction, Routine PCR Procedure and Full-Length 16S rRNA Gene Sequencing
2.5. Genome Sequencing and Data Analysis
2.5.1. Pure Culture DNA Extraction
2.5.2. Whole Genome Sequencing and Gene Annotation
2.6. Accession Number(s)
2.7. Antimicrobial Activity
2.7.1. Bacterial Strains and Growth Conditions
2.7.2. Determination of Inhibitory Properties of LAB Isolates against Foodborne Pathogenic Bacteria and Spoilage Bacteria
Agar Spot Test
Preparations of Cell-Free Supernatant (CFS) and Neutralized CFS (NCFS)
Agar Well Diffusion Assay
Inhibitory Effect during Co-Cultivation
3. Results and Discussion
3.1. UMSC Bacterial Diversity Studies
3.1.1. Analysis of 16S rRNA Gene Fragment Sequences
3.1.2. purR Gene Sequence Analysis
3.1.3. Whole-Genome Sequencing Analysis
3.1.4. Bacterial Library
3.1.5. MALDI-TOF MS and Full-Length 16S rRNA Gene Sequencing Analysis
3.2. UMSC Strains Metabolic and Functional Characteristics
3.2.1. UMSC Genotype–Phenotype Associations
3.2.2. Antimicrobial Activities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Carafa, I.; Stocco, G.; Franceschi, P.; Summer, A.; Tuohy, K.M.; Bittante, G.; Franciosi, E. Evaluation of autochthonous lactic acid bacteria as starter and non-starter cultures for the production of Traditional Mountain cheese. Food Res. Int. 2019, 115, 209–218. [Google Scholar] [CrossRef]
- De Angelis, M.; de Candia, S.; Calasso, M.P.; Faccia, M.; Guinee, T.P.; Simonetti, M.C.; Gobbetti, M. Selection and use of autochthonous multiple strain cultures for the manufacture of high-moisture traditional Mozzarella cheese. Int. J. Food Microbiol. 2008, 125, 123–132. [Google Scholar] [CrossRef]
- De Pasquale, I.; Di Cagno, R.; Buchin, S.; de Angelis, M.; Gobbetti, M. Use of autochthonous mesophilic lactic acid bacteria as starter cultures for making Pecorino Crotonese cheese: Effect on compositional, microbiological and biochemical attributes. Food Res. Int. 2019, 116, 1344–1356. [Google Scholar] [CrossRef]
- Kupiec, B.; Revell, B. Speciality and artisanal cheeses today: The product and the consumer. Br. Food J. 1998, 100, 236–243. [Google Scholar] [CrossRef]
- Antonsson, M.; Molin, G.; Ardö, Y. Lactobacillus strains isolated from Danbo cheese as adjunct cultures in a cheese model system. Int. J. Food Microbiol. 2003, 85, 159–169. [Google Scholar] [CrossRef]
- Bissonnette, F.; Labrie, S.; Deveau, H.; Lamoureux, M.; Moineau, S. Characterization of Mesophilic Mixed Starter Cultures Used for the Manufacture of Aged Cheddar Cheese. J. Dairy Sci. 2000, 83, 620–627. [Google Scholar] [CrossRef]
- Frantzen, C.A.; Kot, W.; Pedersen, T.B.; Ardö, Y.M.; Broadbent, J.R.; Neve, H.; Hansen, L.H.; Dal Bello, F.; Østlie, H.M.; Kleppen, H.P.; et al. Genomic Characterization of Dairy Associated Leuconostoc Species and Diversity of Leuconostocs in Undefined Mixed Mesophilic Starter Cultures. Front. Microbiol. 2017, 8, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carafa, I.; Clementi, F.; Tuohy, K.; Franciosi, E. Microbial evolution of traditional mountain cheese and characterization of early fermentation cocci for selection of autochtonous dairy starter strains. Food Microbiol. 2016, 53, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, B.A.; Cabral, L.; Noronha, M.F.; Baptista, R.C.; Nascimento, H.M.; Sant’Ana, A.S. Amplicon sequencing reveals the bacterial diversity in milk, dairy premises and Serra da Canastra artisanal cheeses produced by three different farms. Food Microbiol. 2020, 89, 1–12. [Google Scholar] [CrossRef]
- Koczura, M.; Martin, B.; Turille, G.; De Marchi, M.; Kreuzer, M.; Berard, J. Milk composition, but not cheese properties, are impaired the day after transhumance to alpine pastures. Int. Dairy J. 2019, 99, 1–9. [Google Scholar] [CrossRef]
- Wouters, J.T.M.; Ayad, E.H.E.; Hugenholtz, J.; Smit, G. Microbes from raw milk for fermented dairy products. Int. Dairy J. 2002, 12, 91–109. [Google Scholar] [CrossRef]
- Taïbi, A.; Dabour, N.; Lamoureux, M.; Roy, D.; LaPointe, G. Evaluation of the genetic polymorphism among Lactococcus lactis subsp. cremoris strains using comparative genomic hybridization and multilocus sequence analysis. Int. J. Food Microbiol. 2010, 144, 20–28. [Google Scholar]
- Smid, E.J.; Kleerebezem, M. Production of aroma compounds in lactic fermentations. Annu. Rev. Food Sci. Technol. 2014, 5, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Buron-Moles, G.; Chailyan, A.; Dolejs, I.; Forster, J.; Mikš, M.H. Uncovering carbohydrate metabolism through a genotype-phenotype association study of 56 lactic acid bacteria genomes. Appl. Microbiol. Biotechnol. 2019, 103, 3135–3152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steensels, J.; Gallone, B.; Voordeckers, K.; Verstrepen, K.J. Domestication of Industrial Microbes. Curr. Biol. 2019, 29, R381–R393. [Google Scholar] [CrossRef]
- Torres Manno, M.; Zuljan, F.; Alarcón, S.; Esteban, L.; Blancato, V.; Espariz, M.; Magni, C. Genetic and phenotypic features defining industrial relevant Lactococcus lactis, L. cremoris and L. lactis biovar. diacetylactis strains. J. Biotechnol. 2018, 282, 25–31. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Huang, J.; Zhou, R. Genomics of lactic acid bacteria: Current status and potential applications. Crit. Rev. Microbiol. 2017, 43, 393–404. [Google Scholar] [CrossRef]
- Bayjanov, J.R.; Starrenburg, M.J.; van der Sijde, M.R.; Siezen, R.J.; van Hijum, S.A. Genotype-phenotype matching analysis of 38 Lactococcus lactis strains using random forest methods. BMC Microbiol. 2013, 13, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ceapa, C.; Lambert, J.; van Limpt, K.; Wels, M.; Smokvina, T.; Knol, J.; Kleerebezem, M. Correlation of Lactobacillus rhamnosus Genotypes and Carbohydrate Utilization Signatures Determined by Phenotype Profiling. Appl. Environ. Microbiol. 2015, 81, 5458–5470. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-H.; Itoh, K.; Kikuchi, E.; Niwa, H.; Fujisawa, T. Identification and Characteristics of Nisin Z-Producing Lactococcus lactis subsp. lactis Isolated from Kimchi. Curr. Microbiol. 2003, 46, 0385–0388. [Google Scholar] [CrossRef]
- Yang, E.; Fan, L.; Jiang, Y.; Doucette, C.; Fillmore, S. Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Express 2012, 2, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor, P.M.; Kuniyoshi, T.M.; Oliveira, R.P.; Hill, C.; Ross, R.P.; Cotter, P.D. Antimicrobials for food and feed; a bacteriocin perspective. Curr. Opin. Biotechnol. 2020, 61, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.D.G.; Brocklehurst, T.F.; Arino, S.; Thuault, D.; Jakobsen, M.; Lange, M.; Farkas, J.; Wimpenny, J.W.T.; van Impe, J.F. Modelling microbial growth in structured foods: Towards a unified approach. Int. J. Food Microbiol. 2002, 73, 275–289. [Google Scholar] [CrossRef]
- Dumarché, C.; Leclercq, S. Method for Preparing a Leaven From Unpasteurized Milk, Leaven Obtained by Means of This Method and Use of This Leaven for Producing a Milk Product. U.S. Patent 20090772755, 12 May 2011. [Google Scholar]
- Leser, T.D.; Amenuvor, J.Z.; Jensen, T.K.; Lindecrona, R.H.; Boye, M.; Møller, K. Culture-Independent Analysis of Gut Bacteria: The Pig Gastrointestinal Tract Microbiota Revisited. Appl. Environ. Microbiol. 2002, 68, 673–690. [Google Scholar] [CrossRef] [Green Version]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G.; et al. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics 2018, 34, 1287–1294. [Google Scholar] [CrossRef]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ 2014, 2, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, 1–22. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G.; et al. MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020, 48, D579–D589. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Ouk Kim, Y.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Van Heel, A.; Jong, A.; Song, C.; Viel, J.; Kok, J.; Kuipers, O. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Bolotin, A.; Wincker, P.; Mauger, S.; Jaillon, O.; Malarme, K.; Weissenbach, J.; Ehrlich, S.D.; Sorokin, A. The Complete Genome Sequence of the Lactic Acid Bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001, 11, 731–753. [Google Scholar] [CrossRef] [Green Version]
- Dubourg, G.; Elsawi, Z.; Raoult, D. Assessment of the in vitro antimicrobial activity of Lactobacillus species for identifying new potential antibiotics. Int. J. Antimicrob. Agents 2015, 46, 590–593. [Google Scholar] [CrossRef]
- Hwanhlem, N.; Ivanova, T.; Haertlé, T.; Jaffrès, E.; Dousset, X. Inhibition of food-spoilage and foodborne pathogenic bacteria by a nisin Z-producing Lactococcus lactis subsp. lactis KT2W2L. LWT—Food Sci. Technol. 2017, 82, 170–175. [Google Scholar] [CrossRef]
- Montel, M.-C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef] [PubMed]
- Turbes, G.; Linscott, T.D.; Tomasino, E.; Waite-Cusic, J.; Lim, J.; Meunier-Goddik, L. Evidence of terroir in milk sourcing and its influence on Cheddar cheese. J. Dairy Sci. 2016, 99, 5093–5103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamei, S.; Hu, Y.O.O.; Olofsson, T.C.; Andersson, A.F.; Forsgren, E.; Vásquez, A. Improvement of identification methods for honeybee specific Lactic Acid Bacteria; future approaches. PLoS ONE 2017, 12, 1–12. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [Green Version]
- Pillidge, C.J.; Sheehy, L.M.; Shihata, A.; Pu, Z.-Y.; Dobos, M.; Powell, I.B. Intragenomic 16S rRNA gene heterogeneity in Lactococcus lactis subsp. cremoris. Int. Dairy J. 2009, 19, 222–227. [Google Scholar] [CrossRef]
- Ndoye, B.; Rasolofo, E.A.; LaPointe, G.; Roy, D. A review of the molecular approaches to investigate the diversity and activity of cheese microbiota. Dairy Sci. Technol. 2011, 91, 495–524. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Zhang, W.; Guo, C.; Yang, X.; Liu, W.; Wu, Y.; Song, Y.; Kwok, L.Y.; Cui, Y.; Menghe, B.; et al. Comparative Genomic Analysis of 45 Type Strains of the Genus Bifidobacterium: A Snapshot of Its Genetic Diversity and Evolution. PLoS ONE 2015, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [Green Version]
- Plummer, E.; Twin, J.; Bulach, D.M.; Garland, S.M.; Tabrizi, S.N. A Comparison of Three Bioinformatics Pipelines for the Analysis of Preterm Gut Microbiota using 16S rRNA Gene Sequencing Data. J. Proteom. Bioinform. 2015, 8, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Song, Y.; Ren, Y.; Qing, Y.; Liu, W.; Sun, Z. Genome-level comparisons provide insight into the phylogeny and metabolic diversity of species within the genus Lactococcus. BMC Microbiol. 2017, 17, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siezen, R.J.; Starrenburg, M.J.C.; Boekhorst, J.; Renckens, B.; Molenaar, D.; van Hylckama Vlieg, J.E. Genome-Scale Genotype-Phenotype Matching of Two Lactococcus lactis Isolates from Plants Identifies Mechanisms of Adaptation to the Plant Niche. Appl. Environ. Microbiol. 2008, 74, 424–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benthin, S.; Nielsen, J.; Villadsen, J. Galactose Expulsion during Lactose Metabolism in Lactococcus lactis subsp. cremoris FD1 Due to Dephosphorylation of Intracellular Galactose 6-Phosphate. Appl. Environ. Microbiol. 1994, 60, 1254–1259. [Google Scholar] [CrossRef] [Green Version]
- Kelly, W.J.; Ward, L.J.H.; Leahy, S.C. Chromosomal Diversity in Lactococcus lactis and the Origin of Dairy Starter Cultures. Genome Biol. Evol. 2010, 2, 729–744. [Google Scholar] [CrossRef] [Green Version]
- Venema, K.; Dost, M.H.; Beun, P.A.; Haandrikman, A.J.; Venema, G.; Kok, J. The genes for secretion and maturation of lactococcins are located on the chromosome of Lactococcus lactis IL1403. Appl. Environ. Microbiol. 1996, 62, 1689–1692. [Google Scholar] [CrossRef] [Green Version]
- Sutherland, J.P.; Bayliss, A.J.; Roberts, T.A. Predictive modelling of growth of Staphylococcus aureus: The effects of temperature, pH and sodium chloride. Int. J. Food Microbiol. 1994, 21, 217–236. [Google Scholar] [CrossRef]
- Alomar, J.; Loubiere, P.; Delbes, C.; Nouaille, S.; Montel, M.C. Effect of Lactococcus garvieae, Lactococcus lactis and Enterococcus faecalis on the behaviour of Staphylococcus aureus in microfiltered milk. Food Microbiol. 2008, 25, 502–508. [Google Scholar] [CrossRef]
- Mufandaedza, J.; Viljoen, B.C.; Feresu, S.B.; Gadaga, T.H. Antimicrobial properties of lactic acid bacteria and yeast-LAB cultures isolated from traditional fermented milk against pathogenic Escherichia coli and Salmonella enteritidis strains. Int. J. Food Microbiol. 2006, 108, 147–152. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.-C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Fath, M.J.; Kolter, R. ABC transporters: Bacterial exporters. Microbiol. Mol. Biol. Rev. 1993, 57, 995–1017. [Google Scholar] [CrossRef]
- Venema, K.; Abee, T.; Haandrikman, A.J.; Leenhouts, K.J.; Kok, J.; Konings, W.N.; Venema, G. Mode of Action of Lactococcin B, a Thiol-Activated Bacteriocin from Lactococcus lactis. Appl. Environ. Microbiol. 1993, 59, 1041–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojic, M.; Strahinic, I.; Fira, D.; Jovcic, B.; Topisirovic, L. Plasmid content and bacteriocin production by five strains of Lactococcus lactis isolated from semi-hard homemade cheese. Can. J. Microbiol. 2006, 52, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Morgan, S.; Ross, R.P.; Hill, C. Bacteriolytic activity caused by the presence of a novel lactococcal plasmid encoding lactococcins A, B, and M. Appl. Environ. Microbiol. 1995, 61, 2995–3001. [Google Scholar] [CrossRef] [Green Version]
- Şahingil, D.; İşleroğlu, H.; Yildirim, Z.; Akçelik, M.; Yildirim, M. Characterization of lactococcin BZ produced by Lactococcus lactis subsp. lactis BZ isolated from boza. Turk. J. Biol. 2011, 35, 21–33. [Google Scholar]

| Growth Conditions | ||
|---|---|---|
| Aerobic atmosphere | 28 °C | BHI agar |
| MRS agar | ||
| M17 agar | ||
| MRS agar + 2% NaCl | ||
| MRS agar + 4% NaCl | ||
| MRS agar + 6.5% NaCl | ||
| BHI agar + 6.5% NaCl | ||
| Microaerophilic atmosphere | 28 °C | BHI agar |
| MRS agar | ||
| M17 agar | ||
| Aerobic atmosphere | 22 °C | BHI agar |
| 37 °C | MRS agar | |
| 44 °C | MRS agar | |
| Strains | Sequencing Technology | Scaffold Number | Genome Size (Mb) | GC % | Predicated Coding Genes | GenBank Accession Number |
|---|---|---|---|---|---|---|
| L. lactis subsp. lactis L1 | Illumina® iSeq | 177 | 2.75 | 34.96 | 2945 | JAATTT000000000 |
| L. lactis subsp. lactis L2 | Illumina® iSeq | 193 | 2.78 | 34.99 | 3003 | JAATTS000000000 |
| L. lactis subsp. lactis L3 | PacBio® RS II | 7 | 2.84 | 35.22 | 3024 | JAATTU000000000 |
| L. lactis subsp. lactis L8 | Illumina® iSeq | 242 | 2.81 | 34.99 | 3036 | JAATTR000000000 |
| L. lactis subsp. lactis L9 | Illumina® iSeq | 229 | 2.80 | 34.98 | 3014 | JAATTQ000000000 |
| L. lactis subsp. lactis L14 | Illumina® iSeq | 178 | 2.78 | 34.98 | 2966 | JAATTP000000000 |
| L. lactis subsp. lactis IL1403 | [40] | [40] | 2.37 | 35.33 | 2473 | AE005176.1 |
| Indicator Strains | Strain | Culture Collection | Growth Medium | Growth Conditions (°C) | Microaerophilic Condition |
|---|---|---|---|---|---|
| Bacillus cereus | CIP 66.24T | CIP 1 | BHI 5 | 30 | No |
| Bacillus subtilis | CIP 52.65T | CIP | BHI | 30 | No |
| Brochothrix thermosphacta | DSMZ 20171T | DSMZ 2 | BHI | 25 | No |
| Carnobacterium maltaromaticum | CIP 103158 | CIP | MRS 6 | 30 | Yes |
| Cronobacter sakazakii | CIP 103183T | CIP | BHI | 30 | No |
| Escherichia coli | DSMZ 1103 | DSMZ | BHI | 37 | No |
| Enterococcus faecalis | CIP 103015 | CIP | BHI | 37 | No |
| Leuconostoc mesenteroides | ATCC 14935 | ATCC 3 | MRS | 30 | Yes |
| Listeria innocua | CIP 80.11 | CIP | BHI | 37 | No |
| Listeria ivanovii | CIP 78.42T | CIP | BHI | 37 | No |
| Listeria monocytogenes | DSMZ 19094 | DSMZ | BHI | 37 | No |
| Listeria monocytogenes | ATCC 35152 | ATCC | BHI | 37 | No |
| Mucor circinelloides | UBOCC-A-112187 | UBOCC 4 | MEA 7 | 25 | No |
| Mucor racemosus | UBOCC-A-111130 | UBOCC | MEA | 25 | No |
| Penicillium solitum | UBOCC-A-111055 | UBOCC | MEA | 25 | No |
| Pseudomonas aeruginosa | ATCC 27853 | ATCC | BHI | 30 | No |
| Pseudomonas fluorescens | CIP 69.13 | CIP | BHI | 30 | No |
| Staphylococcus aureus | DSMZ 13661 | DSMZ | BHI | 37 | No |
| Staphylococcus aureus | CIP 76.25 | CIP | BHI | 37 | No |
| Salmonella ser. Enteritidis | CIP 82.97 | CIP | BHI | 37 | No |
| Salmonella ser. Typhimurium | CIP 104115 | CIP | BHI | 37 | No |
| Yarrowia lipolytica | UBOCC-A-211004 | UBOCC | YPD 8 | 25 | No |
| Yersinia enterocolitica subsp. enterocolitica | DSMZ 4780 | DSMZ | BHI | 26 | No |
| Abundance (%) | |
|---|---|
| ASV_1 | 99.86 |
| ASV_2 | 0.13 |
| ASV_3 | 0.005 |
| ASV_4 | 0.005 |
| ASV_5 | 0.002 |
| Growth Conditions | Number of Collected Isolates | ||
|---|---|---|---|
| Aerobic atmosphere | 28 °C | BHI agar | 128 |
| MRS agar | 192 | ||
| M17 agar | 192 | ||
| MRS agar + 2% NaCl | 96 | ||
| MRS agar + 4% NaCl | 96 | ||
| MRS agar + 6.5% NaCl | 96 | ||
| BHI agar + 6.5% NaCl | 96 | ||
| Microaerophilic atmosphere | 28 °C | BHI agar | 48 |
| MRS agar | 192 | ||
| M17 agar | 192 | ||
| Aerobic atmosphere | 22 °C | BHI agar | 96 |
| 37 °C | MRS agar | 96 | |
| 44 °C | MRS agar | 96 | |
| Total | 1616 | ||
| Carbohydrate | A Group (n = 13) a | B Group (n = 2) b | C Group (n = 2) c | D group (n = 1) d | ATCC 19435 e | ATCC 19257 f | IL1403 h |
|---|---|---|---|---|---|---|---|
| Strain origin | Raw milk | Raw milk | Raw milk | Raw milk | Milk | Cream | Dairy |
| Control | - | - | - | - | - | - | - |
| D-ribose | + | + | + | + | + | - | + |
| L-arabinose | - | - | - | - | - | - | - |
| D-xylose | + | - | + | + | + | - | - |
| D-galactose | + | + | + | + | + | - | + |
| D-glucose | + | + | + | + | + | + | + |
| D-fructose | + | + | + | + | + | + | + |
| D-mannose | + | + | + | + | w g | + | + |
| D-mannitol | - | + | - | + | - | - | - |
| D-sorbitol | - | + | - | - | - | - | - |
| Arbutin | + | + | + | + | + | - | + |
| Salicin | + | + | + | + | + | - | + |
| D-cellobiose | + | + | + | + | + | - | + |
| D-maltose | + | + | + | + | + | - | + |
| D-lactose | + | + | + | + | + | + | w |
| D-melibiose | - | - | - | - | - | - | - |
| D-sucrose | - | + | + | + | - | - | - |
| D-trehalose | + | + | + | + | + | - | + |
| D-raffinose | - | - | - | - | - | - | - |
| Starch | - | - | - | - | - | - | - |
| D-tagatose | - | + | - | - | - | - | - |
| Percentage of carbohydrates (n = 49) utilized per strain (%) | 33 | 45 | 35 | 35 | 29 | 10 | 29 |
| Tested Isolates | S. ser. Typhimurium CIP 104115 | S. aureus DSMZ 13661 | C. maltaromaticum CIP 103158 | S. ser. Enteritidis CIP 82.97 | S. aureus CIP 76.25 | E. faecalis CIP 103015 | L. innocua CIP 80.11 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFS | NCFS | CFS | NCFS | CFS | NCFS | CFS | NCFS | CFS | NCFS | CFS | NCFS | CFS | NCFS | |
| 1 | + | - | - | - | - | - | - | - | + | - | - | - | + | - |
| 3 | + | - | - | - | - | - | - | - | - | - | - | - | + | - |
| 4 | - | - | + | - | - | - | - | - | - | - | + | - | + | - |
| 5 | + | - | - | - | - | - | - | - | - | - | + | - | + | - |
| 6 | + | - | - | - | - | - | - | - | - | - | + | - | + | - |
| 8 | - | - | + | - | - | - | - | - | + | - | + | - | + | - |
| 9 | - | - | + | - | - | - | - | - | - | - | - | - | + | - |
| 10 | - | - | + | - | - | - | - | - | - | - | - | - | + | - |
| 11 | - | - | + | - | - | - | - | - | - | - | + | - | + | - |
| 12 | - | - | + | - | + | - | - | - | - | - | - | - | + | - |
| 13 | - | - | + | - | - | - | - | - | - | - | + | - | + | - |
| 14 | + | - | + | - | - | - | - | - | + | - | + | - | + | + |
| 15 | + | - | + | - | - | - | - | - | + | - | + | - | + | + |
| 16 | - | - | + | - | - | - | - | - | - | - | + | - | + | - |
| 17 | - | - | + | - | - | - | - | - | + | - | - | - | + | - |
| 18 | + | - | + | - | - | - | - | - | - | - | + | - | + | - |
| 20 | + | - | - | - | - | - | - | - | - | - | + | - | + | - |
| 23 | + | - | - | - | - | - | - | - | + | - | - | - | + | - |
| 24 | - | - | - | - | - | - | + | - | + | - | - | - | + | - |
| 25 | + | - | - | - | - | - | - | - | + | - | - | - | + | - |
| 26 | + | - | - | - | - | - | - | - | + | - | - | - | + | - |
| 27 | + | - | - | - | - | - | - | - | + | - | - | - | + | - |
| 30 | - | - | + | - | - | - | - | - | + | - | - | - | + | - |
| 32 | - | - | - | - | - | - | + | - | + | - | - | - | + | - |
| 33 | - | - | + | - | - | - | - | - | + | - | - | - | + | - |
| 34 | + | - | - | - | - | - | - | - | + | - | - | - | + | - |
| Tested CFS | S. ser. Typhimurium CIP 104115 | Lactobacillus Sakei CIP 104494 | S. aureus DSMZ 13661 | L. innocua CIP 80.11 | L. lactis IL1403 d |
|---|---|---|---|---|---|
| L. lactis isolate 14 | 8.0 ± 0.7 a | 12.0 ± 0.7 | 8.0 ± 0.0 | 24.0 ± 0.7 | 13.0 ± 1.4 |
| S. ser. Typhimurium CIP 104115 | /b | / | / | / | / |
| Isolate 14 + S. ser. Typhimurium CIP 104115 | 11.0 ± 0.0 | 14.0 ± 1.4 | ND c | 22.0 ± 1.4 | 15.0 ± 2.12 |
| S. aureus DSMZ 13661 | / | 10.0 ± 2.12 | / | / | / |
| Isolate 14 + S. aureus DSMZ 13661 | ND | 11.0 ± 1.4 | 14.0 ± 0.0 | 18.0 ± 0.7 | 17.0 ± 0.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saltaji, S.; Rué, O.; Sopena, V.; Sablé, S.; Tambadou, F.; Didelot, S.; Chevrot, R. Lactococcus lactis Diversity Revealed by Targeted Amplicon Sequencing of purR Gene, Metabolic Comparisons and Antimicrobial Properties in an Undefined Mixed Starter Culture Used for Soft-Cheese Manufacture. Foods 2020, 9, 622. https://doi.org/10.3390/foods9050622
Saltaji S, Rué O, Sopena V, Sablé S, Tambadou F, Didelot S, Chevrot R. Lactococcus lactis Diversity Revealed by Targeted Amplicon Sequencing of purR Gene, Metabolic Comparisons and Antimicrobial Properties in an Undefined Mixed Starter Culture Used for Soft-Cheese Manufacture. Foods. 2020; 9(5):622. https://doi.org/10.3390/foods9050622
Chicago/Turabian StyleSaltaji, Sabrina, Olivier Rué, Valérie Sopena, Sophie Sablé, Fatoumata Tambadou, Sandrine Didelot, and Romain Chevrot. 2020. "Lactococcus lactis Diversity Revealed by Targeted Amplicon Sequencing of purR Gene, Metabolic Comparisons and Antimicrobial Properties in an Undefined Mixed Starter Culture Used for Soft-Cheese Manufacture" Foods 9, no. 5: 622. https://doi.org/10.3390/foods9050622
APA StyleSaltaji, S., Rué, O., Sopena, V., Sablé, S., Tambadou, F., Didelot, S., & Chevrot, R. (2020). Lactococcus lactis Diversity Revealed by Targeted Amplicon Sequencing of purR Gene, Metabolic Comparisons and Antimicrobial Properties in an Undefined Mixed Starter Culture Used for Soft-Cheese Manufacture. Foods, 9(5), 622. https://doi.org/10.3390/foods9050622