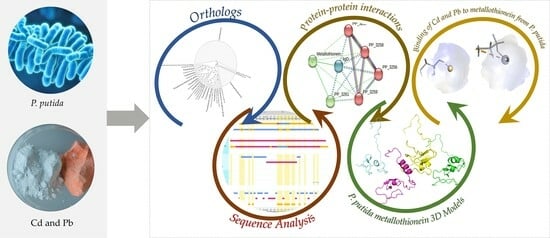
Pseudomonas putida Metallothionein: Structural Analysis and Implications of Sustainable Heavy Metal Detoxification in Madinah
Abstract
:1. Introduction
2. Material and Methods
2.1. Physicochemical Insights: Analysis with EXpasy ProtParam Tool
2.2. Functional Profiling: Unveiling Potential Roles Using VicmPred Algorithm
2.3. Structural Insights: Revealing Architecture via Superfamily 1.75 Tool
2.4. Regulatory Prospects: Post-Translational Modifications Explored with MusiteDeep Tool
2.5. Evolutionary Significance: Functional Annotation through EggNOG 6.0 Database
2.6. Network Exploration: Protein–Protein Interactions Mapped via STRING Database
2.7. Binding Assessment: Capacity Probed using PredictProtein Tool
2.8. Protein Structure Prediction and Validation
2.9. Molecular Docking of Heavy Metals (Lead and Cadmium) with Metallothionein (MT) Modeled Structure and Intramolecular Interaction Analysis
3. Results
3.1. Sequence Analysis
3.2. Ortholog Identification and Analysis of P. putida Metallothionein
3.3. Protein Interaction Profiling of P. putida Metallothionein
3.4. Secondary Structure and Sequence Analysis of P. putida: Insights into Structural Characteristics and Functional Implications
3.5. Three-Dimensional Structure Prediction and Validation
3.6. Molecular Docking and Intramolecular Interactions
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bayumi, T. Impact of Natural and Human Activities on the C roundwater Quality in the Southern Part of AI Madinah Al Munawwarah, Saudi Arabia. Arts Humanit. 2008, 35, 1–21. [Google Scholar]
- Usama, M.; Hussain, N.; Sumrin, A.; Shahbaz, A.; Noor, S.; Bilal, M.; Aleya, L.; Iqbal, H.M.N. Microbial bioremediation strategies with wastewater treatment potentialities—A review. Sci. Total Environ. 2021, 818, 151754. [Google Scholar]
- Kapahi, M.; Sachdeva, S. Bioremediation Options for Heavy Metal Pollution. J. Health Pollut. 2019, 9, 191203. [Google Scholar] [CrossRef] [PubMed]
- Tasleem, M.; Hussein, W.M.; El-Sayed, A.-A.A.A.; Alrehaily, A. An In Silico Bioremediation Study to Identify Essential Residues of Metallothionein Enhancing the Bioaccumulation of Heavy Metals in Pseudomonas aeruginosa. Microorganisms 2023, 11, 2262. [Google Scholar] [CrossRef]
- Maghraby, M.; Nasr, O.; Hamouda, M. Quality assessment of groundwater at south Al Madinah Al Munawarah area, Saudi Arabia. Environ. Earth Sci. 2013, 70, 1525–1538. [Google Scholar] [CrossRef]
- Al Zabadi, H.; Sayeh, G.; Jodeh, S. Environmental exposure assessment of cadmium, lead, copper and zinc in different Palestinian canned foods. Agric. Food Secur. 2018, 7, 50. [Google Scholar] [CrossRef]
- Dahlawi, S.; Al Mulla, A.A.; Saifullah; Salama, K.; Labib, O.A.; Aljassim, M.T.; Akhtar, A.; Asghar, W.; Faraj, T.K.; Khalid, N. Assessment of different heavy metals in cigarette filler and ash from multiple brands retailed in Saudi Arabia. J. King Saud Univ. Sci. 2021, 33, 101521. [Google Scholar] [CrossRef]
- Negi, S.; Batoye, S.; Singh, K.; Waraich, J.S. Environmental Pollution, Its Causes and Impact on Ecosystem; New Frontiers of Nanomaterials in Environmental Science; Springer: Singapore, 2021; pp. 1–22. [Google Scholar]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Mol. Clin. Environ. Toxicol. 2012, 101, 133–164. [Google Scholar] [CrossRef]
- Karri, V.; Schuhmacher, M.; Kumar, V. Heavy metals (Pb, Cd, As and MeHg) as risk factors for cognitive dysfunction: A general review of metal mixture mechanism in brain. Environ. Toxicol. Pharmacol. 2016, 48, 203–213. [Google Scholar] [CrossRef]
- Naranjo, V.I.; Hendricks, M.; Jones, K.S. Lead Toxicity in Children: An Unremitting Public Health Problem. Pediatr. Neurol. 2020, 113, 51–55. [Google Scholar] [CrossRef]
- Schwaba, T.; Bleidorn, W.; Hopwood, C.J.; Gebauer, J.E.; Rentfrow, P.J.; Potter, J.; Gosling, S.D. The impact of childhood lead exposure on adult personality: Evidence from the United States, Europe, and a large-scale natural experiment. Proc. Natl. Acad. Sci. USA 2021, 118, e2020104118. [Google Scholar] [CrossRef]
- Reyes, J.W. Lead exposure and behavior: Effects on antisocial and risky behavior among children and adolescents. Econ. Inq. 2015, 53, 1580–1605. [Google Scholar] [CrossRef]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, A. Cadmium toxicity: Effects on human reproduction and fertility. Rev. Environ. Health 2019, 34, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A. Cadmium & its adverse effects on human health. Ind. J. Med. Res. 2008, 128, 557–564. [Google Scholar]
- Nakazawa, T. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 2002, 4, 782–786. [Google Scholar] [CrossRef]
- Leedjärv, A.; Ivask, A.; Virta, M. Interplay of Different Transporters in the Mediation of Divalent Heavy Metal Resistance in Pseudomonas putida KT2440. J. Bacteriol. 2008, 190, 2680–2689. [Google Scholar] [CrossRef] [PubMed]
- Cánovas, D.; Cases, I.; De Lorenzo, V. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 2003, 5, 1242–1256. [Google Scholar] [CrossRef]
- Isken, S.; Santos, P.M.A.C.; de Bont, J.A.M. Effect of solvent adaptation on the antibiotic resistance in Pseudomonas putida S12. Appl. Microbiol. Biotechnol. 1997, 48, 642–647. [Google Scholar] [CrossRef]
- Gaur, V.K.; Sharma, P.; Gaur, P.; Varjani, S.; Ngo, H.H.; Guo, W.; Chaturvedi, P.; Singhania, R.R. Sustainable mitigation of heavy metals from effluents: Toxicity and fate with recent technological advancements. Bioengineered 2021, 12, 7297–7313. [Google Scholar] [CrossRef]
- Weimer, A.; Kohlstedt, M.; Volke, D.C.; Nikel, P.I.; Wittmann, C. Industrial biotechnology of Pseudomonas putida: Advances and prospects. Appl. Microbiol. Biotechnol. 2020, 104, 7745–7766. [Google Scholar] [CrossRef] [PubMed]
- Kivisaar, M. Narrative of a versatile and adept species Pseudomonas putida. J. Med. Microbiol. 2020, 69, 324–338. [Google Scholar] [CrossRef]
- Maes, S.; De Reu, K.; Van Weyenberg, S.; Lories, B.; Heyndrickx, M.; Steenackers, H. Pseudomonas putida as a potential biocontrol agent against Salmonella Java biofilm formation in the drinking water system of broiler houses. BMC Microbiol. 2020, 20, 373. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvard, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Saha, S.; Raghava, G. VICMpred: An SVM-based Method for the Prediction of Functional Proteins of Gram-negative Bacteria Using Amino Acid Patterns and Composition. Genom. Proteom. Bioinform. 2006, 4, 42–47. [Google Scholar] [CrossRef]
- Gough, J. The SUPERFAMILY database in structural genomics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 1897–1900. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, D.; Yuchi, J.; He, F.; Jiang, Y.; Cai, S.; Li, J.; Xu, D. MusiteDeep: A deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020, 48, W140–W146. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Plaza, A.; Szklarczyk, D.; Botas, J.; Cantalapiedra, C.P.; Giner-Lamia, J.; Mende, D.R.; Kirsch, R.; Rattei, T.; Letunic, I.; Jensen, L.J.; et al. eggNOG 6.0: Enabling comparative genomics across 12 535 organisms. Nucleic Acids Res. 2022, 51, D389–D394. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Rost, B.; Yachdav, G.; Liu, J. The PredictProtein server. Nucleic Acids Res. 2004, 32, W321–W326. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.T.; Ruczinki, I.; Kooperberb, C.; Fox, B.A.; Bystroff, C.; Baker, D. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins Struct. Funct. Bioinform. 1999, 34, 82–95. [Google Scholar] [CrossRef]
- Pieper, U.; Webb, B.M.; Dong, G.Q.; Schneidman-Duhovny, D.; Fan, H.; Kim, S.J.; Khuri, N.; Spill, Y.G.; Weinkam, P.; Hammel, M.; et al. ModBase, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2013, 42, D336–D346. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Uziela, K.; Shu, N.; Wallner, B.; Elofsson, A. ProQ3: Improved model quality assessments using Rosetta energy terms. Sci. Rep. 2016, 6, 33509. [Google Scholar] [CrossRef]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.-C.; Frutiger, S.; Hochstrasser, D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993, 14, 1023–1031. [Google Scholar] [CrossRef]
- Pollastri, G.; McLysaght, A. Porter: A new, accurate server for protein secondary structure prediction. Bioinformatics 2004, 21, 1719–1720. [Google Scholar] [CrossRef] [PubMed]
- Bashford, D.; Karplus, M. pKa’s of ionizable groups in proteins: Atomic detail from a continuum electrostatic model. Biochemistry 1990, 29, 10219–10225. [Google Scholar] [CrossRef]
- Nielsen, J.E.; Gunner, M.; García-Moreno, B.E. The pKa Cooperative: A collaborative effort to advance structure-based calculations of pKa values and electrostatic effects in proteins. Proteins Struct. Funct. Bioinform. 2011, 79, 3249–3259. [Google Scholar] [CrossRef] [PubMed]
- Guruprasad, K.; Reddy, B.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. Des. Sel. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Ikai, A. Thermostability and Aliphatic Index of Globular Proteins. J. Biochem. 1980, 88, 1895–1898. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Emenike, E.C.; Iwuozor, K.O.; Anidiobi, S.U. Heavy Metal Pollution in Aquaculture: Sources, Impacts and Mitigation Techniques. Biol. Trace Element Res. 2021, 200, 4476–4492. [Google Scholar] [CrossRef]
- Kägi, J.H.; Vallee, B.L. Metallothionein: A Cadmium- and Zinc-containing Protein from Equine Renal Cortex. J. Biol. Chem. 1960, 235, 3460–3465. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, J.; Oh, B.; Kimm, K.; Koh, I. Prediction of phosphorylation sites using SVMs. Bioinformatics 2004, 20, 3179–3184. [Google Scholar] [CrossRef] [PubMed]
- Ning, W.; Jiang, P.; Guo, Y.; Wang, C.; Tan, X.; Zhang, W.; Peng, D.; Xue, Y. GPS-Palm: A deep learning-based graphic presentation system for the prediction of S-palmitoylation sites in proteins. Briefings Bioinform. 2020, 22, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zheng, Y.; Li, H.; Luo, X.; He, Z.; Cao, S.; Shi, Y.; Zhao, Q.; Xue, Y.; Zuo, Z.; et al. GPS-Lipid: A robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 2016, 6, 28249. [Google Scholar] [CrossRef]
- Liu, Z.; Cao, J.; Gao, X.; Ma, Q.; Ren, J.; Xue, Y. GPS-CCD: A Novel Computational Program for the Prediction of Calpain Cleavage Sites. PLoS ONE 2011, 6, e19001. [Google Scholar] [CrossRef]
- Zhao, Q.; Xie, Y.; Zheng, Y.; Jiang, S.; Liu, W.; Mu, W.; Liu, Z.; Zhao, Y.; Xue, Y.; Ren, J. GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014, 42, W325–W330. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Kumari, S.; Rath, S.; Priyadarshanee, M.; Das, S. Diversity, structure and regulation of microbial metallothionein: Metal resistance and possible applications in sequestration of toxic metals. Metallomics 2020, 12, 1637–1655. [Google Scholar] [CrossRef]
- Tiquia-Arashiro, S.M. Lead absorption mechanisms in bacteria as strategies for lead bioremediation. Appl. Microbiol. Biotechnol. 2018, 102, 5437–5444. [Google Scholar] [CrossRef]
- Murzin, A.G. Structural classification of proteins: New superfamilies. Curr. Opin. Struct. Biol. 1996, 6, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Kägi, J.H.R.; Kojima, Y.; Kissling, M.M.; Lerch, K. Metallothionein: An Exceptional Metal Thiolate Protein; Wiley: Hoboken, NJ, USA, 1980; pp. 223–237. [Google Scholar] [CrossRef]
- Klaassen, C.D.; Cagen, S.Z. Metallothionein as a trap for reactive organic intermediates. In Biological Reactive Intermediates—II: Chemical Mechanisms and Biological Effects Part A; Springer: Berlin/Heidelberg, Germany, 2012; pp. 633–646. [Google Scholar]
- Aslebagh, R.; Wormwood, K.L.; Channaveerappa, D.; Wetie, A.G.N.; Woods, A.G.; Darie, C.C. Identification of Posttranslational Modifications (PTMs) of Proteins by Mass Spectrometry. In Advancements of Mass Spectrometry in Biomedical Research; Advances in Experimental Medicine and Biology; Woods, A., Darie, C., Eds.; Springer: Cham, Switzerland, 2019; Volume 1140, pp. 199–224. [Google Scholar] [CrossRef]
- Su, M.-G.; Weng, J.T.-Y.; Hsu, J.B.-K.; Huang, K.-Y.; Chi, Y.-H.; Lee, T.-Y. Investigation and identification of functional post-translational modification sites associated with drug binding and protein-protein interactions. BMC Syst. Biol. 2017, 11, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, M.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.Z.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, gkw937. [Google Scholar] [CrossRef]
- Samson, S.L.-A.; Gedamu, L. Molecular analyses of metallothionein gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 1997, 59, 257–288. [Google Scholar]
- Daneshvar, E.; Zarrinmehr, M.J.; Kousha, M.; Hashtjin, A.M.; Saratale, G.D.; Maiti, A.; Vithanage, M.; Bhatnagar, A. Hexavalent chromium removal from water by microalgal-based materials: Adsorption, desorption and recovery studies. Bioresour. Technol. 2019, 293, 122064. [Google Scholar] [CrossRef]
- Andrews, G.K. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem. Pharmacol. 1999, 59, 95–104. [Google Scholar] [CrossRef]
- Rascio, N.; Navari-Izzo, F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Logan, D.T.; Danielsson, J.; Oliveberg, M. Exposing the distinctive modular behavior of β-strands and α-helices in folded proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 28775–28783. [Google Scholar] [CrossRef] [PubMed]
- Nilges, M. Homology Modeling. In Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine; Springer: Berlin/Heidelberg, Germany, 2006; pp. 814–817. [Google Scholar]
- Shin, W.-H.; Kang, X.; Zhang, J.; Kihara, D. Prediction of Local Quality of Protein Structure Models Considering Spatial Neighbors in Graphical Models. Sci. Rep. 2017, 7, 40629. [Google Scholar] [CrossRef]
- Sanders, J.T.; Golloshi, R.; Das, P.; Xu, Y.; Terry, P.H.; Nash, D.G.; Dekker, J.; McCord, R.P. Loops, topologically associating domains, compartments, and territories are elastic and robust to dramatic nuclear volume swelling. Sci. Rep. 2022, 12, 4721. [Google Scholar] [CrossRef]
- Grennan, A.K. Metallothioneins, a Diverse Protein Family. Plant Physiol. 2011, 155, 1750–1751. [Google Scholar] [CrossRef] [PubMed]
- Ngu, T.T.; Stillman, M.J. Metal-binding mechanisms in metallothioneins. Dalton Trans. 2009, 28, 5425–5433. [Google Scholar] [CrossRef] [PubMed]







| Model Validation Tool | Phyre2 | Robetta | ModWeb | SwissModel |
|---|---|---|---|---|
| Residues built | 1–74 | 1–74 | 1–73 | 1–73 |
| ProQ3 | 0.383 | 0.467 | 0.423 | 0.000 |
| Ramachandran Plot Summary | ||||
| Most favored | 84.1% | 95.4% | 87.1% | 75.8% |
| Additionally allowed | 12.7% | 4.6% | 11.3% | 21.0% |
| Generously allowed | 3.2% | 0.0% | 1.6% | 3.2% |
| Disallowed | 0.0% | 0.0% | 0.0% | 0.0% |
| Close Contacts and Deviations from Ideal Geometry | ||||
| Number of close contacts (within 2.2 Å) | 0 | 0 | 0 | 0 |
| RMS deviation for bond angles | 1.9° | 2.2° | 2.3° | 2.2° |
| RMS deviation for bond lengths | 0.020 Å | 0.016 Å | 0.020 Å | 0.016 Å |
| Global quality scores | ||||
| Verify3D | −6.10 | −5.78 | −6.90 | −6.74 |
| ProsaII (-ve) | −0.37 | −0.37 | 1.03 | −0.79 |
| Procheck (phi-psi) 3 | −0.43 | 0.51 | −0.75 | −3.03 |
| Procheck (all) 3 | 0.24 | 1.18 | −0.59 | −2.90 |
| MolProbity Clashscore | −19.42 | 1.09 | 1.53 | 1.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tasleem, M.; El-Sayed, A.-A.A.A.; Hussein, W.M.; Alrehaily, A. Pseudomonas putida Metallothionein: Structural Analysis and Implications of Sustainable Heavy Metal Detoxification in Madinah. Toxics 2023, 11, 864. https://doi.org/10.3390/toxics11100864
Tasleem M, El-Sayed A-AAA, Hussein WM, Alrehaily A. Pseudomonas putida Metallothionein: Structural Analysis and Implications of Sustainable Heavy Metal Detoxification in Madinah. Toxics. 2023; 11(10):864. https://doi.org/10.3390/toxics11100864
Chicago/Turabian StyleTasleem, Munazzah, Abdel-Aziz A. A. El-Sayed, Wesam M. Hussein, and Abdulwahed Alrehaily. 2023. "Pseudomonas putida Metallothionein: Structural Analysis and Implications of Sustainable Heavy Metal Detoxification in Madinah" Toxics 11, no. 10: 864. https://doi.org/10.3390/toxics11100864
APA StyleTasleem, M., El-Sayed, A.-A. A. A., Hussein, W. M., & Alrehaily, A. (2023). Pseudomonas putida Metallothionein: Structural Analysis and Implications of Sustainable Heavy Metal Detoxification in Madinah. Toxics, 11(10), 864. https://doi.org/10.3390/toxics11100864