Transcriptomic Analysis of Diethylstilbestrol in Daphnia Magna: Energy Metabolism and Growth Inhibition
Abstract
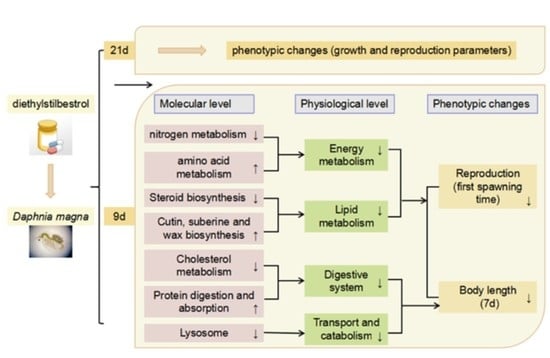
:1. Introduction
2. Materials and Methods
2.1. D. magna Culture Maintenance
2.2. Chemicals
2.3. Acute and Chornic Toxicity Test
2.4. RNA Sequencing
2.5. RNA-Seq Data Analyses
2.6. Quantitative Real-Time PCR
2.7. Statistical Analyses
3. Results
3.1. Effects of Acute and Chronic Toxicity Test of DES
3.2. Transcriptome Analysis
3.2.1. Differentially Expressed Genes
3.2.2. Analyses of Gene Ontology and Functional Pathway
4. Discussion
4.1. Genes Related to Energy Metabolism
4.2. Genes Related to Lipid Metabolism
4.3. Genes Related to Digestive System
4.4. Genes Related to Transport and Catabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carpenter, S.R.; Kitchell, J.F.; Hodgson, J.R.; Cochran, P.A.; Elser, J.J.; Elser, M.M.; Lodge, D.M.; Kretchmer, D.; He, X.; von Ende, C.N. Regulation of Lake Primary Productivity by Food Web Structure. Ecology 1987, 68, 1863–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tessier, A.J.; Leibold, M.A.; Tsao, J. A Fundamental Trade-Off in Resource Exploitation by Daphnia and Consequences to Plankton Communities. Ecology 2000, 81, 826. [Google Scholar] [CrossRef]
- Akbar, S.; Gu, L.; Sun, Y.; Zhang, L.; Lyu, K.; Huang, Y.; Yang, Z. Understanding host-microbiome-environment interactions: Insights from Daphnia as a model organism. Sci. Total. Environ. 2022, 808, 152093. [Google Scholar] [CrossRef] [PubMed]
- Toyota, K.; McNabb, N.A.; Spyropoulos, D.D.; Iguchi, T.; Kohno, S. Toxic effects of chemical dispersant Corexit 9500 on water flea Daphnia magna. J. Appl. Toxicol. 2017, 37, 201–206. [Google Scholar] [CrossRef]
- Bownik, A.; Kowalczyk, M.; Bańczerowski, J. Lambda-cyhalothrin affects swimming activity and physiological responses of Daphnia magna. Chemosphere 2019, 216, 805–811. [Google Scholar] [CrossRef]
- Gowler, C.D.; Rogalski, M.A.; Shaw, C.L.; Hunsberger, K.K.; Duffy, M.A. Density, parasitism, and sexual reproduction are strongly correlated in lake Daphnia populations. Ecol. Evol. 2021, 11, 10446–10456. [Google Scholar] [CrossRef]
- Brockmeier, E.K.; Hodges, G.; Hutchinson, T.H.; Butler, E.; Hecker, M.; Tollefsen, K.E.; Garcia-Reyero, N.; Kille, P.; Becker, D.; Chipman, K.; et al. The Role of Omics in the Application of Adverse Outcome Pathways for Chemical Risk Assessment. Toxicol. Sci. 2017, 158, 252–262. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Ito, S.; Nguyen, H.T.; Yamamoto, K.; Tanoue, R.; Kunisue, T.; Iwata, H. Effects of prenatal exposure to triclosan on the liver transcriptome in chicken embryos. Toxicol. Appl. Pharmacol. 2018, 347, 23–32. [Google Scholar] [CrossRef]
- Fuertes, I.; Jordão, R.; Piña, B.; Barata, C. Time-dependent transcriptomic responses of Daphnia magna exposed to metabolic disruptors that enhanced storage lipid accumulation. Environ. Pollut. 2019, 249, 99–108. [Google Scholar] [CrossRef]
- Zheng, Y.; Yuan, J.; Gu, Z.; Yang, G.; Li, T.; Chen, J. Transcriptome alterations in female Daphnia (Daphnia magna) exposed to 17β-estradiol. Environ. Pollut. 2020, 261, 114208. [Google Scholar] [CrossRef]
- Hunt, P.A.; Sathyanarayana, S.; Fowler, P.A.; Trasande, L. Female Reproductive Disorders, Diseases, and Costs of Exposure to Endocrine Disrupting Chemicals in the European Union. J. Clin. Endocrinol. Metab. 2016, 101, 1562–1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Ding, H.; Yu, X.; Shi, X.; Sun, A.; Li, D.; Zhao, J. Characterization and application of molecularly imprinted polymer-coated quantum dots for sensitive fluorescent determination of diethylstilbestrol in water samples. Talanta 2019, 197, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Reed, C.E.; Fenton, S.E. Exposure to diethylstilbestrol during sensitive life stages: A legacy of heritable health effects. Birth Defects Res. Part C Embryo Today Rev. 2013, 99, 134–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Sun, J.; Jia, H.; Zhang, Y.; Pang, L.; He, L.; Chai, T. A disposable paper-based sample clean-up slides for the sensitive determination of trace diethylstilbestrol residues in aquatic products. Microchem. J. 2019, 151, 104243. [Google Scholar] [CrossRef]
- Król, J.; Pobłocki, W.; Bockenheimer, T.; Hliwa, P. Effect of diethylstilbestrol (DES) and 17 β-estradiol (E2) on growth, survival and histological structure of the internal organs in juvenile European catfish Silurus glanis (L.). Aquac. Int. 2014, 22, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-C.; Kuo, H.-W.; Ding, W.-H. Determination of estrogenic compounds in wastewater using liquid chromatography–tandem mass spectrometry with electrospray and atmospheric pressure photoionization following desalting extraction. Chemosphere 2009, 74, 508–514. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Zhang, H.; Luo, Z.; Yan, C. Occurrence, distribution, and seasonal variation of estrogenic compounds and antibiotic residues in Jiulongjiang River, South China. Environ. Sci. Pollut. Res. 2012, 19, 1392–1404. [Google Scholar] [CrossRef]
- Nikov, G.N.; Eshete, M.; Rajnarayanan, R.V.; Alworth, W.L. Interactions of synthetic estrogens with human estrogen receptors. J. Endocrinol. 2001, 170, 137–145. [Google Scholar] [CrossRef] [Green Version]
- Shang, G.; Xue, J.; Li, M.; Hu, H.-Y.; Lu, Y. Estrogen receptor affinity chromatography: A new method for characterization of novel estrogenic disinfection by-products. Chemosphere 2014, 104, 251–257. [Google Scholar] [CrossRef]
- Baldwin, W.S.; Milam, D.L.; Leblanc, G.A. Physiological and biochemical perturbations inDaphnia magnafollowing exposure to the model environmental estrogen diethylstilbestrol. Environ. Toxicol. Chem. 1995, 14, 945–952. [Google Scholar] [CrossRef]
- Brennan, S.J.; Brougham, C.A.; Roche, J.J.; Fogarty, A.M. Multi-generational effects of four selected environmental oestrogens on Daphnia magna. Chemosphere 2006, 64, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Kobayashi, K.; Kato, Y.; Oda, S.; Abe, R.; Tatarazako, N.; Iguchi, T. Transcriptome profiling in crustaceans as a tool for ecotoxicogenomics. Cell Biol. Toxicol. 2008, 24, 641–647. [Google Scholar] [CrossRef] [PubMed]
- David, R.M.; Dakic, V.; Williams, T.D.; Winter, M.J.; Chipman, J.K. Transcriptional responses in neonate and adult Daphnia magna in relation to relative susceptibility to genotoxicants. Aquat. Toxicol. 2011, 104, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Palma, P.; Palma, V.; Fernandes, R.; Bohn, A.; Soares, A.; Barbosa, I. Embryo-toxic effects of environmental concentrations of chlorpyrifos on the crustacean Daphnia magna. Ecotoxicol. Environ. Saf. 2009, 72, 1714–1718. [Google Scholar] [CrossRef]
- OECD. Test No. 202: Daphnia sp. Acute Immobilisation Test, OECD Guidelines for the Testing of Chemicals, Section 2; OECD Publishing: Paris, France, 2004. [Google Scholar] [CrossRef] [Green Version]
- OECD. Test No. 211: Daphnia magna Reproduction Test, OECD Guidelines for the Testing of Chemicals, Section 2; OECD Publishing: Paris, France, 2012. [Google Scholar] [CrossRef] [Green Version]
- Federico, A.; Serra, A.; Ha, M.K.; Kohonen, P.; Choi, J.-S.; Liampa, I.; Nymark, P.; Sanabria, N.; Cattelani, L.; Fratello, M.; et al. Transcriptomics in Toxicogenomics, Part II: Preprocessing and Differential Expression Analysis for High Quality Data. Nanomaterials 2020, 10, 903. [Google Scholar] [CrossRef]
- Kowal, K.; Tkaczyk, A.; Pierzchała, M.; Bownik, A.; Ślaska, B. Identification of Mitochondrial DNA (NUMTs) in the Nuclear Genome of Daphnia magna. Int. J. Mol. Sci. 2020, 21, 8725. [Google Scholar] [CrossRef]
- Supuran, C.T.; Simone, G.D. Carbonic Anhydrases as Biocatalysts: From Theory to Medical and Industrial Applications; Elsevier: Amsterdam, The Netherlands, 2015; ISBN 9780444632586. [Google Scholar] [CrossRef]
- Hu, L.; Diez-Fernandez, C.; Ruefenacht, V.; Hismi, B.Ö.; Ünal, Ö.; Soyucen, E.; Çoker, M.; Bayraktar, B.T.; Gunduz, M.; Kiykim, E.; et al. Recurrence of carbamoyl phosphate synthetase 1 (CPS1) deficiency in Turkish patients: Characterization of a founder mutation by use of recombinant CPS1 from insect cells expression. Mol. Genet. Metab. 2014, 113, 267–273. [Google Scholar] [CrossRef]
- Shi, J.; Jiang, Y.; Jiang, Z.; Wang, X.; Wang, X.; Zhang, S.; Han, P.; Yang, C. Enzymatic conversion of carbon dioxide. Chem. Soc. Rev. 2015, 44, 5981–6000. [Google Scholar] [CrossRef]
- Tu, Y.; Xie, S.; Han, D.; Yang, Y.; Jin, J.; Zhu, X. Dietary arginine requirement for gibel carp (Carassis auratus gibelio var. CAS III) reduces with fish size from 50g to 150g associated with modulation of genes involved in TOR signaling pathway. Aquaculture 2015, 449, 37–47. [Google Scholar] [CrossRef]
- Suzuki, A.; Iwata, J. Amino acid metabolism and autophagy in skeletal development and homeostasis. Bone 2021, 146, 115881. [Google Scholar] [CrossRef]
- Kim, Y.T.; Song, Y.H.; Churchich, J.E. Recombinant brain 4-aminobutyrate aminotransferases overexpression, purification, and identification of Lys-330 at the active site. Biochim. et Biophys. Acta (BBA)-Protein Struct. Mol. Enzym. 1997, 1337, 248–256. [Google Scholar] [CrossRef]
- Fernández-Veledo, S.; Ceperuelo-Mallafré, V.; Vendrell, J. Rethinking succinate: An unexpected hormone-like metabolite in energy homeostasis. Trends Endocrinol. Metab. 2021, 32, 680–692. [Google Scholar] [CrossRef]
- Basoli, V.; Li, Z.; Traweger, A.; Sanchez-Antequera, Y.; Plank, C.; Rip, J.; Alini, M.; Grad, S. Effect of nanoparticle based mrna delivery on modulation of inflammation in an osteochondral inflammation model. Osteoarthr. Cartil. 2021, 29, S13. [Google Scholar] [CrossRef]
- Ballantine, J.A.; Roberts, J.C.; Morris, R.J. The sterols of crustaceans: Decapods (Sub-order macrura). Comp. Biochem. Physiol. Part B Comp. Biochem. 1980, 67, 75–79. [Google Scholar] [CrossRef]
- Bard, M.; A Bruner, D.; A Pierson, C.; Lees, N.D.; Biermann, B.; Frye, L.; Koegel, C.; Barbuch, R. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc. Natl. Acad. Sci. USA 1996, 93, 186–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, D.M.; Chen, O.S.; Li, L.; Kaplan, J.; Bhuiyan, S.A.; Natarajan, S.K.; Bard, M.; Cox, J.E. Altered sterol metabolism in budding yeast affects mitochondrial iron–sulfur (Fe-S) cluster synthesis. J. Biol. Chem. 2018, 293, 10782–10795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suchodolski, J.; Muraszko, J.; Bernat, P.; Krasowska, A. A Crucial Role for Ergosterol in Plasma Membrane Composition, Localisation, and Activity of Cdr1p and H+-ATPase in Candida albicans. Microorganisms 2019, 7, 378. [Google Scholar] [CrossRef] [Green Version]
- Hofvander, P.; Doan, T.T.; Hamberg, M. A prokaryotic acyl-CoA reductase performing reduction of fatty acyl-CoA to fatty alcohol. FEBS Lett. 2011, 585, 3538–3543. [Google Scholar] [CrossRef] [Green Version]
- Sargent, J.; Gatten, R.; McIntosh, R. Biosynthesis of wax esters in cell-free preparations of euchaeta norvegica. Comp. Biochem. Physiol. Part B Comp. Biochem. 1974, 47, 217–227. [Google Scholar] [CrossRef]
- Bauermeister, A.; Sargent, J. Wax esters: Major metabolites in the marine environment. Trends Biochem. Sci. 1979, 4, 209–211. [Google Scholar] [CrossRef]
- Norlin, M.; Wikvall, K. Enzymes in the conversion of cholesterol into bile acids. Curr. Mol. Med. 2007, 7, 199–218. [Google Scholar] [CrossRef]
- Dubland, J.A.; Francis, G.A. Lysosomal acid lipase: At the crossroads of normal and atherogenic cholesterol metabolism. Front. Cell Dev. Biol. 2015, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, K.; Balch, W.E. NPC1/NPC2 function as a tag team duo to mobilize cholesterol. Proc. Natl. Acad. Sci. USA 2008, 105, 15223–15224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxfield, F.R.; Menon, A.K. Intracellular sterol transport and distribution. Curr. Opin. Cell Biol. 2006, 18, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Xie, A.-J.; Mai, C.-T.; Zhu, Y.-Z.; Liu, X.-C.; Xie, Y. Bile acids as regulatory molecules and potential targets in metabolic diseases. Life Sci. 2021, 287, 120152. [Google Scholar] [CrossRef]
- Whitcomb, D.C.; Lowe, M.E. Human Pancreatic Digestive Enzymes. Dig. Dis. Sci. 2007, 52, 1–17. [Google Scholar] [CrossRef]
- Koenig, A.; Mueller, C.; Hasel, C.; Adler, G.; Menke, A. Collagen Type I Induces Disruption of E-Cadherin–Mediated Cell-Cell Contacts and Promotes Proliferation of Pancreatic Carcinoma Cells. Cancer Res. 2006, 66, 4662–4671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Liu, R.; Shan, Y.; Sun, C. Marine bacterial exopolysaccharide EPS11 inhibits migration and invasion of liver cancer cells by directly targeting collagen I. J. Biol. Chem. 2021, 297, 101133. [Google Scholar] [CrossRef]
- Emanuel, R.; Sergin, I.; Bhattacharya, S.; Turner, J.N.; Epelman, S.; Settembre, C.; Diwan, A.; Ballabio, A.; Razani, B. Induction of Lysosomal Biogenesis in Atherosclerotic Macrophages Can Rescue Lipid-Induced Lysosomal Dysfunction and Downstream Sequelae. Arter. Thromb. Vasc. Biol. 2014, 34, 1942–1952. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.-Q.; Li, Q.; Hu, Z.-H.; Liu, H.-C.; Rao, C.-L.; Zhang, M.-J.; Xia, Y.-P.; Deng, L.; Mao, X.-H.; Fang, Y. MicroRNA-146a inhibits autophagy to maintain the intracellular survival of Burkholderia pseudomallei by targeting LIPA. Microb. Pathog. 2021, 158, 104969. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.; Futerman, A.H. Lysosomal storage disorders: Old diseases, present and future challenges. Pediatr. Endocrinol. Rev. 2013, 11 (Suppl. S1), 59–63. [Google Scholar]
- Lei, K.; Lin, C.-Y.; Zhu, Y.; Chen, W.; Pan, H.-Y.; Sun, Z.; Sweetman, A.; Zhang, Q.; He, M.-C. Estrogens in municipal wastewater and receiving waters in the Beijing-Tianjin-Hebei region, China: Occurrence and risk assessment of mixtures. J. Hazard. Mater. 2020, 389, 121891. [Google Scholar] [CrossRef] [PubMed]




| Pathways | Category | Up-Genes | Down-Genes |
|---|---|---|---|
| SC vs. L | |||
| Steroid biosynthesis | Lipid metabolism | - | meso1, erg25 |
| Terpenoid backbone biosynthesis | Metabolism of terpenoids and polyketides | - | pdss1 |
| Nitrogen metabolism | Energy metabolism | - | ca |
| Cell adhesion molecules (CAMs) | Signaling molecules and interaction | - | cntnap2 |
| SC vs. M | |||
| Cholesterol metabolism | Digestive system | - | npc2 |
| Lysosome | Transport and catabolism | hgsnat | npc |
| Cutin, suberine, and wax biosynthesis | Lipid metabolism | far | - |
| Glycosaminoglycan degradation | Glycan biosynthesis and metabolism | hgsnat | - |
| SC vs. H | |||
| Cholesterol metabolism | Digestive system | lal | npc1, npc2 |
| Lysosome | Transport and catabolism | lipa | npc1, npc2 |
| Cutin, suberine, and wax biosynthesis | Lipid metabolism | far | - |
| Alanine, aspartate, and glutamate metabolism | Amino acid metabolism | abat | - |
| Protein digestion and absorption | Digestive system | prss1_2_3, col1a | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Zhao, Q.; Guo, J.; Li, X.; Song, J. Transcriptomic Analysis of Diethylstilbestrol in Daphnia Magna: Energy Metabolism and Growth Inhibition. Toxics 2023, 11, 197. https://doi.org/10.3390/toxics11020197
Li Q, Zhao Q, Guo J, Li X, Song J. Transcriptomic Analysis of Diethylstilbestrol in Daphnia Magna: Energy Metabolism and Growth Inhibition. Toxics. 2023; 11(2):197. https://doi.org/10.3390/toxics11020197
Chicago/Turabian StyleLi, Qi, Qian Zhao, Jiahua Guo, Xi Li, and Jinxi Song. 2023. "Transcriptomic Analysis of Diethylstilbestrol in Daphnia Magna: Energy Metabolism and Growth Inhibition" Toxics 11, no. 2: 197. https://doi.org/10.3390/toxics11020197
APA StyleLi, Q., Zhao, Q., Guo, J., Li, X., & Song, J. (2023). Transcriptomic Analysis of Diethylstilbestrol in Daphnia Magna: Energy Metabolism and Growth Inhibition. Toxics, 11(2), 197. https://doi.org/10.3390/toxics11020197