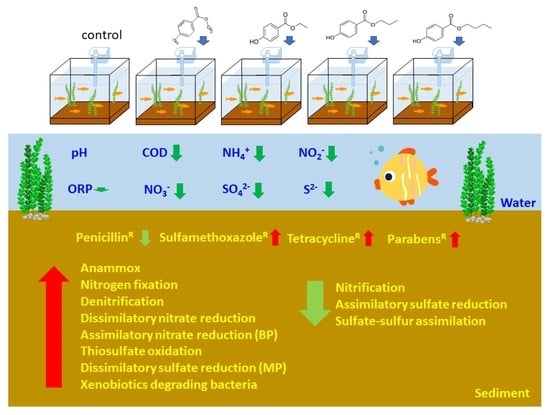
Parabens Increase Sulfamethoxazole-, Tetracycline- and Paraben-Resistant Bacteria and Reshape the Nitrogen/Sulfur Cycle-Associated Microbial Communities in Freshwater River Sediments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Experimental Design
2.3. Bacterial Culture and Plate Count
2.4. Analysis of Chemical Compositions in Water
2.5. HPLC Analysis of Residual Parabens in Water
2.6. DNA Extraction, 16S Amplicon Preparation and NGS
2.7. Microbiome Data Analysis
3. Results
3.1. Increased Tetracycline-, Sulfamethoxazole- and Paraben-Resistant Microbes in Sediment
3.2. Analysis of Chemical Compositions and Oxidation-Reduction Potential (ORP) in Water
3.3. Analysis of Microbial Community Compositions
3.4. Microbial Community Associated with the Nitrogen Cycle
3.5. Microbial Community Associated with the Sulfur Cycle
3.6. Microbial Communities Associated with Xenobiotics Degradation and Pathogenic Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Janjua, N.R.; Mortensen, G.K.; Andersson, A.M.; Kongshoj, B.; Skakkebaek, N.E.; Wulf, H.C. Systemic uptake of diethyl phthalate, dibutyl phthalate, and butyl paraben following whole-body topical application and reproductive and thyroid hormone levels in humans. Environ. Sci. Technol. 2007, 41, 5564–5570. [Google Scholar] [CrossRef] [PubMed]
- Andersen, F.A. Final amended report on the safety assessment of Methylparaben, Ethylparaben, Propylparaben, Isopropylparaben, Butylparaben, Isobutylparaben, and Benzylparaben as used in cosmetic products. Int. J. Toxicol. 2008, 27 (Suppl. S4), 1–82. [Google Scholar]
- Nowak, K.; Jabłońska, E.; Ratajczak-Wrona, W. Controversy around parabens: Alternative strategies for preservative use in cosmetics and personal care products. Environ. Res. 2021, 198, 110488. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.; Ratajczak-Wrona, W.; Górska, M.; Jabłońska, E. Parabens and their effects on the endocrine system. Mol. Cell Endocrinol. 2018, 474, 238–251. [Google Scholar] [CrossRef]
- Karwacka, A.; Zamkowska, D.; Radwan, M.; Jurewicz, J. Exposure to modern, widespread environmental endocrine disrupting chemicals and their effect on the reproductive potential of women: An overview of current epidemiological evidence. Hum. Fertil. 2019, 22, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Dash, P.R.; Darbre, P.D. Exposure to parabens at the concentration of maximal proliferative response increases migratory and invasive activity of human breast cancer cells in vitro. J. Appl. Toxicol. 2014, 34, 1051–1059. [Google Scholar] [CrossRef]
- Błędzka, D.; Gromadzińska, J.; Wąsowicz, W. Parabens. From environmental studies to human health. Environ. Int. 2014, 67, 27–42. [Google Scholar] [CrossRef]
- Yamamoto, H.; Tamura, I.; Hirata, Y.; Kato, J.; Kagota, K.; Katsuki, S.; Yamamoto, A.; Kagami, Y.; Tatarazako, N. Aquatic toxicity and ecological risk assessment of seven parabens: Individual and additive approach. Sci. Total Environ. 2011, 410–411, 102–111. [Google Scholar] [CrossRef]
- Kasprzyk-Hordern, B.; Dinsdale, R.M.; Guwy, A.J. The occurrence of pharmaceuticals, personal care products, endocrine disruptors and illicit drugs in surface water in South Wales, UK. Water Res. 2008, 42, 3498–3518. [Google Scholar] [CrossRef]
- González-Mariño, I.; Quintana, J.B.; Rodríguez, I.; Cela, R. Evaluation of the occurrence and biodegradation of parabens and halogenated by-products in wastewater by accurate-mass liquid chromatography-quadrupole-time-of-flight-mass spectrometry (LC-QTOF-MS). Water Res. 2011, 45, 6770–6780. [Google Scholar] [CrossRef]
- Lu, J.; Li, H.; Luo, Z.; Lin, H.; Yang, Z. Occurrence, distribution, and environmental risk of four categories of personal care products in the Xiangjiang River, China. Environ. Sci. Pollut. Res. Int. 2018, 25, 27524–27534. [Google Scholar] [CrossRef] [PubMed]
- Haman, C.; Dauchy, X.; Rosin, C.; Munoz, J.F. Occurrence, fate and behavior of parabens in aquatic environments: A review. Water Res. 2015, 68, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhao, J.; Xi, N.; Guo, W.; Sun, J. Parabens and their metabolite in surface water and sediment from the Yellow River and the Huai River in Henan Province: Spatial distribution, seasonal variation and risk assessment. Ecotoxicol. Environ. Saf. 2019, 172, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Lee, H.K.; Moon, H.B. Contamination and spatial distribution of parabens, their metabolites and antimicrobials in sediment from Korean coastal waters. Ecotoxicol. Environ. Saf. 2019, 180, 185–191. [Google Scholar] [CrossRef]
- Amin, A.; Chauhan, S.; Dare, M.; Bansal, A.K. Degradation of parabens by Pseudomonas beteli and Burkholderia latens. Eur. J. Pharm. Biopharm. 2010, 75, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Onuche, P.; Okibe, F.; Ajibola, V. Biodegradation of sodium dodecyl sulphate and methyl paraben in shampoo and hair dressing salon waste by bacteria from sewage treatment sludge. Am. Chem. Sci. 2016, 14, 1–9. [Google Scholar] [CrossRef]
- Bolujoko, N.B.; Unuabonah, E.I.; Alfred, M.O.; Ogunlaja, A.; Ogunlaja, O.O.; Omorogie, M.O.; Olukanni, O.D. Toxicity and removal of parabens from water: A critical review. Sci. Total Environ. 2021, 792, 148092. [Google Scholar] [CrossRef]
- Wei, F.; Mortimer, M.; Cheng, H.; Sang, N.; Guo, L.H. Parabens as chemicals of emerging concern in the environment and humans: A review. Sci. Total Environ. 2021, 778, 146150. [Google Scholar] [CrossRef]
- Alam, M.Z.; Aqil, F.; Ahmad, I.; Ahmad, S. Incidence and transferability of antibiotic resistance in the enteric bacteria isolated from hospital wastewater. Braz. J. Microbiol. 2014, 44, 799–806. [Google Scholar] [CrossRef]
- Choi, C.; Ham, H.J.; Kwon, D.; Kim, J.; Cheon, D.S.; Min, K.; Cho, W.S.; Chung, H.K.; Jung, T.; Jung, K.; et al. Antimicrobial susceptibility of pathogenic Escherichia coli isolated from pigs in Korea. J. Vet. Med. Sci. 2002, 64, 71–73. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Roemhild, R.; Andersson, D.I. Mechanisms and therapeutic potential of collateral sensitivity to antibiotics. PLoS Pathog. 2021, 17, e1009172. [Google Scholar] [CrossRef] [PubMed]
- Corona, F.; Blanco, P.; Alcalde-Rico, M.; Hernando-Amado, S.; Lira, F.; Bernardini, A.; Sánchez, M.B.; Martínez, J.L. The analysis of the antibiotic resistome offers new opportunities for therapeutic intervention. Future Med. Chem. 2016, 8, 1133–1151. [Google Scholar] [CrossRef] [PubMed]
- Rosenkilde, C.E.H.; Munck, C.; Porse, A.; Linkevicius, M.; Andersson, D.I.; Sommer, M.O.A. Collateral sensitivity constrains resistance evolution of the CTX-M-15 β-lactamase. Nat. Commun. 2019, 10, 618. [Google Scholar] [CrossRef]
- Gonzales, P.R.; Pesesky, M.W.; Bouley, R.; Ballard, A.; Biddy, B.A.; Suckow, M.A.; Wolter, W.R.; Schroeder, V.A.; Burnham, C.A.; Mobashery, S.; et al. Synergistic, collaterally sensitive β-lactam combinations suppress resistance in MRSA. Nat. Chem. Biol. 2015, 11, 855–861. [Google Scholar] [CrossRef]
- Herencias, C.; Rodríguez-Beltrán, J.; León-Sampedro, R.; Alonso-Del Valle, A.; Palkovičová, J.; Cantón, R.; San Millán, Á. Collateral sensitivity associated with antibiotic resistance plasmids. Elife 2021, 10, e65130. [Google Scholar] [CrossRef]
- Pantosti, A.; Sanchini, A.; Monaco, M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007, 2, 323–334. [Google Scholar] [CrossRef]
- Leal, L.H.; Vieno, N.; Temmink, H.; Zeeman, G.; Buisman, C.J. Occurrence of xenobiotics in gray water and removal in three biological treatment systems. Environ. Sci. Technol. 2010, 44, 6835–6842. [Google Scholar] [CrossRef]
- Valkova, N.; Lépine, F.; Valeanu, L.; Dupont, M.; Labrie, L.; Bisaillon, J.G.; Beaudet, R.; Shareck, F.; Villemur, R. Hydrolysis of 4-hydroxybenzoic acid esters (parabens) and their aerobic transformation into phenol by the resistant Enterobacter cloacae strain EM. Appl. Environ. Microbiol. 2001, 67, 2404–2409. [Google Scholar] [CrossRef]
- Nguyen, V.H.; Phan Thi, L.A.; Chandana, P.S.; Do, H.T.; Pham, T.H.; Lee, T.; Nguyen, T.D.; Le Phuoc, C.; Huong, P.T. The degradation of paraben preservatives: Recent progress and sustainable approaches toward photocatalysis. Chemosphere 2021, 276, 130163. [Google Scholar] [CrossRef]
- Leoni, B.; Patelli, M.; Soler, V.; Nava, V. Ammonium Transformation in 14 Lakes along a Trophic Gradient. Water 2018, 10, 265. [Google Scholar] [CrossRef]
- Collos, Y.; Harrison, P.J. Acclimation and toxicity of high ammonium concentrations to unicellular algae. Mar. Pollut. Bull. 2014, 80, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Zekker, I.; Rikmann, E.; Tenno, T.; Vabamäe, P.; Kroon, K.; Loorits, L.; Saluste, A.; Tenno, T. Effect of concentration on anammox nitrogen removal rate in a moving bed biofilm reactor. Environ. Technol. 2012, 33, 2263–2271. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Zhang, S.; Li, S.; Zhang, L.; Wang, G.; Zhang, L.; Wang, J.; Li, Z. The cycle of nitrogen in river systems: Sources, transformation, and flux. Environ. Sci. Process Impacts. 2018, 20, 863–891. [Google Scholar] [CrossRef]
- He, Z.; Feng, Y.; Zhang, S.; Wang, X.; Wu, S.; Pan, X. Oxygenic denitrification for nitrogen removal with less greenhouse gas emissions: Microbiology and potential applications. Sci. Total Environ. 2018, 621, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Chan-Pacheco, C.R.; Valenzuela, E.I.; Cervantes, F.J.; Quijano, G. Novel biotechnologies for nitrogen removal and their coupling with gas emissions abatement in wastewater treatment facilities. Sci. Total Environ. 2021, 797, 149228. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Song, B.; Phillips, R.L.; Chang, J.; Song, M.J. Ecological and physiological implications of nitrogen oxide reduction pathways on greenhouse gas emissions in agroecosystems. FEMS Microbiol. Ecol. 2019, 95, fiz066. [Google Scholar] [CrossRef]
- Song, B.; Lisa, J.A.; Tobias, C.R. Linking DNRA community structure and activity in a shallow lagoonal estuarine system. Front. Microbiol. 2014, 5, 460. [Google Scholar] [CrossRef]
- Hardison, A.K.; Algar, C.K.; Giblin, A.E.; Rich, J.J. Influence of organic carbon and nitrate loading on partitioning between dissimilatory nitrate reduction to ammonium (DNRA) and N2 production. Geochim. Cosmochim. Acta 2015, 164, 146–160. [Google Scholar] [CrossRef]
- Giblin, A.; Tobias, C.; Song, B.; Weston, N.; Banta, G.T.; Rivera-Monroy, V.H. The importance of dissimilatory nitrate reduction to ammonium (DNRA) in the nitrogen cycle of coastal ecosystems. Oceanography 2013, 26, 124–131. [Google Scholar] [CrossRef]
- Keeler, B.L.; Gourevitch, J.D.; Polasky, S.; Isbell, F.; Tessum, C.W.; Hill, J.D.; Marshall, J.D. The social costs of nitrogen. Sci. Adv. 2016, 2, e1600219. [Google Scholar] [CrossRef] [PubMed]
- Rios-Del Toro, E.E.; Valenzuela, E.I.; Lopez-Lozano, N.E.; Cortes-Martinez, M.G.; Sanchez-Rodriguez, M.A.; Calvario-Martinez, O.; Sanchez-Carrillo, S.; Cervantes, F.J. Anaerobic ammonium oxidation linked to sulfate and ferric iron reduction fuels nitrogen loss in marine sediments. Biodegradation 2018, 29, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Y.; Xie, G.J.; Xing, D.F.; Liu, B.F.; Ding, J.; Cao, G.L.; Ren, N.Q. Sulfate dependent ammonium oxidation: A microbial process linked nitrogen with sulfur cycle and potential application. Environ. Res. 2021, 192, 110282. [Google Scholar] [CrossRef]
- Dang, C.; Liu, S.; Chen, Q.; Sun, W.; Zhong, H.; Hu, J.; Liang, E.; Ni, J. Response of microbial nitrogen transformation processes to antibiotic stress in a drinking water reservoir. Sci. Total Environ. 2021, 797, 149119. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-W.; Lee, W.-C. Parabens Increase Sulfamethoxazole-, Tetracycline- and Paraben-Resistant Bacteria and Reshape the Nitrogen/Sulfur Cycle-Associated Microbial Communities in Freshwater River Sediments. Toxics 2023, 11, 387. https://doi.org/10.3390/toxics11040387
Yang C-W, Lee W-C. Parabens Increase Sulfamethoxazole-, Tetracycline- and Paraben-Resistant Bacteria and Reshape the Nitrogen/Sulfur Cycle-Associated Microbial Communities in Freshwater River Sediments. Toxics. 2023; 11(4):387. https://doi.org/10.3390/toxics11040387
Chicago/Turabian StyleYang, Chu-Wen, and Wei-Chen Lee. 2023. "Parabens Increase Sulfamethoxazole-, Tetracycline- and Paraben-Resistant Bacteria and Reshape the Nitrogen/Sulfur Cycle-Associated Microbial Communities in Freshwater River Sediments" Toxics 11, no. 4: 387. https://doi.org/10.3390/toxics11040387
APA StyleYang, C. -W., & Lee, W. -C. (2023). Parabens Increase Sulfamethoxazole-, Tetracycline- and Paraben-Resistant Bacteria and Reshape the Nitrogen/Sulfur Cycle-Associated Microbial Communities in Freshwater River Sediments. Toxics, 11(4), 387. https://doi.org/10.3390/toxics11040387