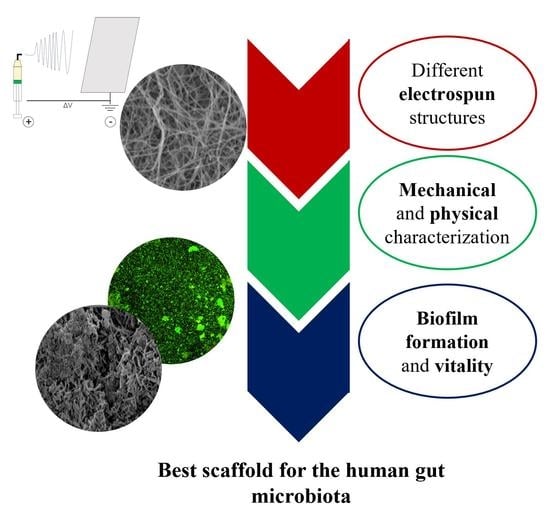
Study of the Adhesion of the Human Gut Microbiota on Electrospun Structures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Electrospun Structures Fabrication
2.2. Contact Angle Measurement and Mechanical/Physical Characterization
2.3. Microbiota Preparation from Stool Samples
2.4. Microbial Growth on Electrospun Structures
2.5. Biofilm Biomass Measurement
2.6. Live/Dead and Scanning Electron Microscopy Imaging
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garrett, T.R.; Bhakoo, M.; Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Biagini, F.; Calvigioni, M.; Lapomarda, A.; Vecchione, A.; Magliaro, C.; De Maria, C.; Montemurro, F.; Celandroni, F.; Mazzantini, D.; Mattioli-Belmonte, M.; et al. A novel 3D in vitro model of the human gut microbiota. Sci. Rep. 2020, 10, 21499–21510. [Google Scholar] [CrossRef]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, 1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS. Suppl. 2013, 121, 1–51. [Google Scholar] [CrossRef]
- Achinas, S.; Charalampogiannis, N.; Euverink, G.J.W. A brief recap of microbial adhesion and biofilms. Appl. Sci. 2019, 9, 2801. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Guégan, C.; Garderes, J.; Le Pennec, G.; Gaillard, F.; Fay, F.; Linossier, I.; Herry, J.M.; Fontaine, M.N.B.; Réhel, K.V. Alteration of bacterial adhesion induced by the substrate stiffness. Colloids Surf. B Biointerfaces 2014, 114, 193–200. [Google Scholar] [CrossRef]
- Hou, S.; Gu, H.; Smith, C.; Ren, D. Microtopographic patterns affect escherichia coli biofilm formation on poly(dimethylsiloxane) surfaces. Langmuir 2011, 27, 2686–2691. [Google Scholar] [CrossRef]
- Chen, D.C.; Avansino, J.R.; Agopian, V.G.; Hoagland, V.D.; Woolman, J.D.; Pan, S.; Ratner, B.D.; Stelzner, M. Comparison of Polyester Scaffolds for Bioengineered Intestinal Mucosa. Cells Tissues Organs 2006, 184, 154–165. [Google Scholar] [CrossRef]
- Yuan, Y.; Hays, M.P.; Hardwidge, P.R.; Kim, J. Surface characteristics influencing bacterial adhesion to polymeric substrates. RSC Adv. 2017, 7, 14254–14261. [Google Scholar] [CrossRef] [Green Version]
- Song, F.; Ren, D. Stiffness of cross-linked poly(dimethylsiloxane) affects bacterial adhesion and antibiotic susceptibility of attached cells. Langmuir 2014, 30, 10354–10362. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Koo, H.; Ren, D. Effects of material properties on bacterial adhesion and biofilm formation. J. Dent. Res. 2015, 94, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Dantas, L.C.D.M.; Da Silva-Neto, J.P.; Dantas, T.S.; Naves, L.Z.; Das Neves, F.D.; Da Mota, A.S. Bacterial adhesion and surface roughness for different clinical techniques for acrylic polymethyl methacrylate. Int. J. Dent. 2016, 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, R.J.; Webb, H.K.; Truong, V.K.; Hasan, J.; Ivanova, E.P. Surface topographical factors influencing bacterial attachment. Adv. Colloid Interface Sci. 2012, 179–182, 142–149. [Google Scholar] [CrossRef]
- Ge, X.; Leng, Y.; Lu, X.; Ren, F.; Wang, K.; Ding, Y.; Yang, M. Bacterial responses to periodic micropillar array. J. Biomed. Mater. Res. Part A 2015, 103, 384–396. [Google Scholar] [CrossRef]
- Yang, M.; Ding, Y.; Ge, X.; Leng, Y. Control of bacterial adhesion and growth on honeycomb-like patterned surfaces. Colloids Surf. B Biointerfaces 2015, 135, 549–555. [Google Scholar] [CrossRef]
- Jun, I.; Han, H.S.; Edwards, J.R.; Jeon, H. Electrospun Fibrous Scaffolds for Tissue Engineering: Viewpoints on Architecture and Fabrication. Int. J. Mol. Sci. 2018, 19, 745. [Google Scholar] [CrossRef] [Green Version]
- Rahmati, M.; Mills, D.K.; Urbanska, A.M.; Saeb, M.R.; Venugopal, J.R.; Ramakrishna, S.; Mozafari, M. Electrospinning for tissue engineering applications. Prog. Mater. Sci. 2021, 117, 100721. [Google Scholar] [CrossRef]
- De Cesare, F.; Di Mattia, E.; Zussman, E.; Macagnano, A. A study on the dependence of bacteria adhesion on the polymer nanofibre diameter. Environ. Sci. Nano 2019, 6, 778–797. [Google Scholar] [CrossRef]
- Herrmann, M.; Vaudaux, P.; Pittet, D.; Auckenthaler, R.; Lew, P.; Schumacher-Perdreau, F.; Peters, G.; Waldvogel, F. Fibronectin, fibrinogen, and laminin act as mediators of adherence of clinical staphylococcal isolates to foreign material. J. Infect. Dis. 1988, 158, 693–701. [Google Scholar] [CrossRef]
- Herrmann, M.; Suchard, S.J.; Boxer, L.A.; Waldvogel, F.A.; Lew, P.D. Thrombospondin binds to Staphylococcus aureus and promotes staphylococcal adherence to surfaces. Infect. Immun. 1991, 59, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulsson, M.; Kober, M.; Freij-Larsson, C.; Stollenwerk, M.; Wesslén, B.; Ljungh, Å. Adhesion of staphylococci to chemically modified and native polymers, and the influence of preadsorbed fibronectin, vitronectin and fibrinogen. Biomaterials 1993, 14, 845–853. [Google Scholar] [CrossRef]
- Celebioglu, H.; Svensson, B. Dietary Nutrients, Proteomes, and Adhesion of Probiotic Lactobacilli to Mucin and Host Epithelial Cells. Microorganisms 2018, 6, 90. [Google Scholar] [CrossRef] [Green Version]
- Van Tassell, M.L.; Miller, M.J. Lactobacillus Adhesion to Mucus. Nutrients 2011, 3, 613–636. [Google Scholar] [CrossRef] [Green Version]
- Boekhorst, J.; Helmer, Q.; Kleerebezem, M.; Siezen, R. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 2006, 152, 273–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okutan, N.; Terzi, P.; Altay, F. Affecting parameters on electrospinning process and characterization of electrospun gelatin nanofibers. Food Hydrocoll. 2014, 39, 19–26. [Google Scholar] [CrossRef]
- Ginestra, P.; Ceretti, E.; Fiorentino, A. Electrospinning of Poly-caprolactone for Scaffold Manufacturing: Experimental Investigation on the Process Parameters Influence. Procedia CIRP 2016, 49, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Shi, L.; Zhang, X.; Liu, K.; Ullah, I.; Cheng, P. Electrospinning of polycaprolactone nanofibers using H2O as benign additive in polycaprolactone/glacial acetic acid solution. J. Appl. Polym. Sci. 2018, 135, 45578. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajilić-Stojanović, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Wijman, J.G.E.; de Leeuw, P.P.L.A.; Moezelaar, R.; Zwietering, M.H.; Abee, T. Air-liquid interface biofilms of Bacillus cereus: Formation, sporulation, and dispersion. Appl. Environ. Microbiol. 2007, 73, 1481–1488. [Google Scholar] [CrossRef] [Green Version]
- Crémet, L.; Corvec, S.; Batard, E.; Auger, M.; Lopez, I.; Pagniez, F.; Dauvergne, S.; Caroff, N. Comparison of three methods to study biofilm formation by clinical strains of Escherichia coli. Diagn. Microbiol. Infect. Dis. 2013, 75, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Francolini, I.; Vuotto, C.; Piozzi, A.; Donelli, G. Antifouling and antimicrobial biomaterials: An overview. APMIS 2017, 125, 392–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, W.; Zhai, Z.; Gao, C. Adaptive antibacterial biomaterial surfaces and their applications. Mater. Today Bio 2019, 2, 100017. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.C.; Coia, H.G.; Karl, J.P.; Pantoja-Feliciano, I.G.; Zachos, N.C.; Racicot, K. Intestinal in vitro and ex vivo models to study host-microbiome interactions and acute stressors. Front. Physiol. 2018, 9, 1584. [Google Scholar] [CrossRef] [Green Version]
- Trujillo-de Santiago, G.; Lobo-Zegers, M.J.; Montes-Fonseca, S.L.; Zhang, Y.S.; Alvarez, M.M. Gut-microbiota-on-a-chip: An enabling field for physiological research. Microphysiol. Syst. 2018, 1. [Google Scholar] [CrossRef]
- Kargar, M.; Wang, J.; Nain, A.S.; Behkam, B. Controlling bacterial adhesion to surfaces using topographical cues: A study of the interaction of Pseudomonas aeruginosa with nanofiber-textured surfaces. Soft Matter 2012, 8, 10254–10259. [Google Scholar] [CrossRef]
- Lichter, J.A.; Thompson, M.T.; Delgadillo, M.; Nishikawa, T.; Rubner, M.F.; Van Vliet, K.J. Substrata mechanical stiffness can regulate adhesion of viable bacteria. Biomacromolecules 2008, 9, 1571–1578. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Guan, A.; Isayeva, I.; Vorvolakos, K.; Das, S.; Li, Z.; Phillips, K.S. Interactions of Staphylococcus aureus with ultrasoft hydrogel biomaterials. Biomaterials 2016, 95, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Verhorstert, K.; Guler, Z.; de Boer, L.; Riool, M.; Roovers, J.; Zaat, S. In Vitro Bacterial Adhesion and Biofilm Formation on Fully Absorbable Poly-4-hydroxybutyrate and Nonabsorbable Polypropylene Pelvic Floor Implants. ACS Appl. Mater. Interfaces 2020, 12, 53646–53653. [Google Scholar] [CrossRef]
- De-la-Pinta, I.; Cobos, M.; Ibarretxe, J.; Montoya, E.; Eraso, E.; Guraya, T.; Quindós, G. Effect of biomaterials hydrophobicity and roughness on biofilm development. J. Mater. Sci. Mater. Med. 2019, 30, 1–11. [Google Scholar] [CrossRef]
- Røder, H.L.; Sørensen, S.J.; Burmølle, M. Studying Bacterial Multispecies Biofilms: Where to Start? Trends Microbiol. 2016, 24, 503–513. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, D.A.; Jeffers, F.; Parker, M.L.; Vibert-Vallet, A.; Bongaerts, R.J.; Roos, S.; Walter, J.; Juge, N. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology 2010, 156, 3368–3378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickett, M.J.; Greenwood, J.R.; Harvey3, S.M. Tests for Detecting Degradation of Gelatin: Comparison of Five Methods. J. Clin. Microbiol. 1991, 29, 2322–2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breure, A.M.; van Andel, J.G. Hydrolysis and acidogenic fermentation of a protein, gelatin, in an anaerobic continuous culture. Appl. Microbiol. Biotechnol. 1984, 20, 40–45. [Google Scholar] [CrossRef]



| Contact Angle | |
| Gelatin | 28.6 ± 0.5° |
| PCL | 109.4 ± 5.2° |
| Gelatin + mucin | 17.5 ± 2.0° |
| PCL + mucin | 31.7 ± 5.2° |
| Elastic Modulus | |
| Gelatin (dry) | 23.8 ± 2.6 MPa |
| Gelatin (wet) | 0.199 ± 0.04 MPa |
| PCL (dry) | 2.1 ± 0.5 MPa |
| Diameter of Fibers | |
| Gelatin | 0.32 ± 0.03 µm |
| PCL | 0.38 ± 0.1 µm |
| Porosity | |
| Gelatin | 31.5 ± 1.8% |
| PCL | 23 ± 4.7% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biagini, F.; Calvigioni, M.; De Maria, C.; Magliaro, C.; Montemurro, F.; Mazzantini, D.; Celandroni, F.; Mattioli-Belmonte, M.; Ghelardi, E.; Vozzi, G. Study of the Adhesion of the Human Gut Microbiota on Electrospun Structures. Bioengineering 2022, 9, 96. https://doi.org/10.3390/bioengineering9030096
Biagini F, Calvigioni M, De Maria C, Magliaro C, Montemurro F, Mazzantini D, Celandroni F, Mattioli-Belmonte M, Ghelardi E, Vozzi G. Study of the Adhesion of the Human Gut Microbiota on Electrospun Structures. Bioengineering. 2022; 9(3):96. https://doi.org/10.3390/bioengineering9030096
Chicago/Turabian StyleBiagini, Francesco, Marco Calvigioni, Carmelo De Maria, Chiara Magliaro, Francesca Montemurro, Diletta Mazzantini, Francesco Celandroni, Monica Mattioli-Belmonte, Emilia Ghelardi, and Giovanni Vozzi. 2022. "Study of the Adhesion of the Human Gut Microbiota on Electrospun Structures" Bioengineering 9, no. 3: 96. https://doi.org/10.3390/bioengineering9030096
APA StyleBiagini, F., Calvigioni, M., De Maria, C., Magliaro, C., Montemurro, F., Mazzantini, D., Celandroni, F., Mattioli-Belmonte, M., Ghelardi, E., & Vozzi, G. (2022). Study of the Adhesion of the Human Gut Microbiota on Electrospun Structures. Bioengineering, 9(3), 96. https://doi.org/10.3390/bioengineering9030096