A Comprehensive Insight into Fungal Enzymes: Structure, Classification, and Their Role in Mankind’s Challenges
Abstract
:1. Introduction
2. Fungal Enzymes Structure and Function
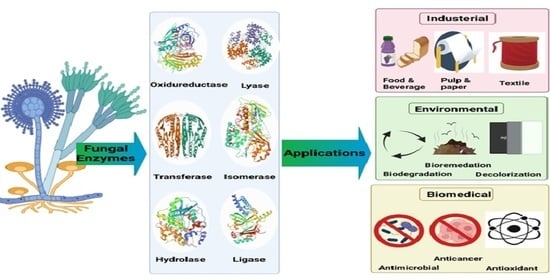
3. Fungal Enzyme Nomenclature and Classifications
3.1. Oxidoreductases (EC 1)
3.2. Transferases (EC 2)
3.3. Hydrolases (EC 3)
3.4. Lyases (EC 4)
3.5. Isomerases (EC 5)
3.6. Ligases (EC 6)
4. Application of Fungal Enzymes
4.1. Industrial Applications
4.1.1. Food and Beverage
4.1.2. Pulp and Paper
4.1.3. Textile
4.2. Environmental Applications
Bioremediation
4.3. Biomedical Applications
4.3.1. Antimicrobial
4.3.2. Anticancer
4.3.3. Antioxidant
5. Toward Safe, Sustainable Production and Application of Fungal Enzymes
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Guerrand, D. Economics of food and feed enzymes: Status and prospectives. In Enzymes in Human and Animal Nutrition: Principles and Perspectives; Nunes, C.S., Kumar, V., Eds.; Academic Press: London, UK, 2018; pp. 487–514. [Google Scholar]
- Singh, R.; Singh, T.; Pandey, A. Microbial enzymes—An overview. In Advances in Enzyme Technology; Singh, R.S., Singhania, R.R., Pandey, A., Larroche, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–40. [Google Scholar]
- Kango, N.; Jana, U.K.; Choukade, R. Fungal Enzymes: Sources and Biotechnological Applications. In Advancing Frontiers in Mycology & Mycotechnology: Basic and Applied Aspects of Fungi; Satyanarayana, T., Deshmukh, S.K., Deshpande, M.V., Eds.; Springer: Singapore, 2019; pp. 515–538. ISBN 9789811393488. [Google Scholar]
- Kumla, J.; Suwannarach, N.; Sujarit, K.; Penkhrue, W.; Kakumyan, P.; Jatuwong, K.; Vadthanarat, S.; Lumyong, S. Cultivation of Mushrooms and Their Lignocellulolytic Enzyme Production Through the Utilization of Agro-Industrial Waste. Molecules 2020, 25, 2811. [Google Scholar] [CrossRef]
- Da Luz, J.M.R.; Nunes, M.D.; Paes, S.A.; Torres, D.P.; da Silva, M.D.C.S.; Kasuya, M.C.M. Lignocellulolytic enzyme production of Pleurotus ostreatus growth in agroindustrial wastes. Braz. J. Microbiol. 2012, 43, 1508–1515. [Google Scholar] [CrossRef]
- Fen, L.; Xuwei, Z.; Nanyi, L.; Puyu, Z.; Shuang, Z.; Xue, Z.; Pengju, L.; Qichao, Z.; Haiping, L. Screening of Lignocellulose-Degrading Superior Mushroom Strains and Determination of Their CMCase and Laccase Activity. Sci. World J. 2014, 2014, 763108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, F.F.G.; Junior, S.B.; Hantao, L.W.; Augusto, F.; Sato, H.H. Acrylamide mitigation in French fries using native L-asparaginase from Aspergillus oryzae CCT 3940. LWT—Food Sci. Technol. 2017, 76, 222–229. [Google Scholar] [CrossRef]
- Meghavarnam, A.K.; Janakiraman, S. Evaluation of acrylamide reduction potential of l-asparaginase from Fusarium culmorum (ASP-87) in starchy products. LWT 2018, 89, 32–37. [Google Scholar] [CrossRef]
- Pedreschi, F.; Mariotti, S.; Granby, K.; Risum, J. Acrylamide reduction in potato chips by using commercial asparaginase in combination with conventional blanching. LWT—Food Sci. Technol. 2011, 44, 1473–1476. [Google Scholar] [CrossRef]
- Renzetti, S.; Courtin, C.; Delcour, J.; Arendt, E. Oxidative and proteolytic enzyme preparations as promising improvers for oat bread formulations: Rheological, biochemical and microstructural background. Food Chem. 2010, 119, 1465–1473. [Google Scholar] [CrossRef]
- Pal, A.; Khanum, F. Efficacy of xylanase purified from Aspergillus niger DFR-5 alone and in combination with pectinase and cellulase to improve yield and clarity of pineapple juice. J. Food Sci. Technol. 2011, 48, 560–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salim, D.; Anwar, Z.; Zafar, M.; Anjum, A.; Bhatti, K.H.; Irshad, M. Pectinolytic cocktail: Induced yield and its exploitation for lignocellulosic materials saccharification and fruit juice clarification. Food Biosci. 2018, 22, 154–164. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Tu, T.; Zhang, D.; Ma, R.; You, S.; Wang, X.; Yao, B.; Luo, H.; Xu, B. Two acidic, thermophilic GH28 polygalacturonases from Talaromyces leycettanus JCM 12802 with application potentials for grape juice clarification. Food Chem. 2017, 237, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, D.; Camarero, S.; Romero, J.; Martínez, M.J.; Martínez, A.T. Integrating laccase–mediator treatment into an industrial-type sequence for totally chlorine-free bleaching of eucalypt kraft pulp. J. Chem. Technol. Biotechnol. 2006, 81, 1159–1165. [Google Scholar] [CrossRef]
- Garcia-Ubasart, J.; Esteban, A.; Vila, C.; Roncero, M.B.; Colom, J.F.; Vidal, T. Enzymatic treatments of pulp using laccase and hydrophobic compounds. Bioresour. Technol. 2011, 102, 2799–2803. [Google Scholar] [CrossRef] [PubMed]
- Campioni, T.S.; de Jesus Moreira, L.; Moretto, E.; Nunes, N.S.S.; de Oliva Neto, P. Biobleaching of Kraft pulp using fungal xylanases produced from sugarcane straw and the subsequent decrease of chlorine consumption. Biomass Bioenergy 2019, 121, 22–27. [Google Scholar] [CrossRef]
- Nathan, V.K.; Rani, M.E.; Gunaseeli, R.; Kannan, N.D. Enhanced biobleaching efficacy and heavy metal remediation through enzyme mediated lab-scale paper pulp deinking process. J. Clean. Prod. 2018, 203, 926–932. [Google Scholar] [CrossRef]
- Londoño-Hernandez, L.; Ruiz, H.A.; Ramírez, T.C.; Ascacio, J.A.; Rodríguez-Herrera, R.; Aguilar, C.N. Biocatalysis and Agricultural Biotechnology Fungal detoxification of coffee pulp by solid-state fermentation. Biocatal. Agric. Biotechnol. 2020, 23, 101467. [Google Scholar] [CrossRef]
- Sigoillot, C.; Camarero, S.; Vidal, T.; Record, E.; Asther, M.; Pérez-Boada, M.; Martínez, M.J.; Sigoillot, J.-C.; Asther, M.; Colom, J.F.; et al. Comparison of different fungal enzymes for bleaching high-quality paper pulps. J. Biotechnol. 2005, 115, 333–343. [Google Scholar] [CrossRef]
- Wang, H.; Kaur, G.; Pensupa, N.; Uisan, K.; Du, C.; Yang, X.; Lin, C.S.K. Textile waste valorization using submerged filamentous fungal fermentation. Process. Saf. Environ. Prot. 2018, 118, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Du, C.; Leu, S.-Y.; Jing, H.; Li, X.; Lin, C.S.K. Valorisation of textile waste by fungal solid state fermentation: An example of circular waste-based biorefinery. Resour. Conserv. Recycl. 2018, 129, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Abd El Aty, A.A.; Saleh, S.A.; Eid, B.M.; Ibrahim, N.A.; Mostafa, F.A. Thermodynamics characterization and potential textile applications of Trichoderma longibrachiatum KT693225 xylanase. Biocatal. Agric. Biotechnol. 2018, 14, 129–137. [Google Scholar] [CrossRef]
- Aggarwal, R.; Dutta, T.; Sheikh, J. Extraction of amylase from the microorganism isolated from textile mill effluent vis a vis desizing of cotton. Sustain. Chem. Pharm. 2019, 14, 100178. [Google Scholar] [CrossRef]
- Aggarwal, R.; Dutta, T.; Sheikh, J. Extraction of pectinase from Candida isolated from textile mill effluent and its application in bio-scouring of cotton. Sustain. Chem. Pharm. 2020, 17, 100291. [Google Scholar] [CrossRef]
- Farooq, A.; Ali, S.; Abbas, N.; Fatima, G.A.; Ashraf, M.A. Comparative performance evaluation of conventional bleaching and enzymatic bleaching with glucose oxidase on knitted cotton fabric. J. Clean. Prod. 2013, 42, 167–171. [Google Scholar] [CrossRef]
- Manai, I.; Miladi, B.; El Mselmi, A.; Smaali, I.; Ben Hassen, A.; Hamdi, M.; Bouallagui, H. Industrial textile effluent decolourization in stirred and static batch cultures of a new fungal strain Chaetomium globosum IMA1 KJ472923. J. Environ. Manag. 2016, 170, 8–14. [Google Scholar] [CrossRef]
- Linhartová, L.; Michalíková, K.; Šrédlová, K.; Cajthaml, T. Biodegradability of Dental Care Antimicrobial Agents. Molecules 2020, 25, 400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vantamuri, A.B.; Kaliwal, B.B. Purification and characterization of laccase from Marasmius species BBKAV79 and effective decolorization of selected textile dyes. 3 Biotech 2016, 6, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchut-Mikolajczyk, O.; Kwapisz, E.; Wieczorek, D.; Antczak, T.Z. Biodegradation of diesel oil hydrocarbons enhanced with Mucor circinelloides enzyme preparation. Int. Biodeterior. Biodegrad. 2015, 104, 142–148. [Google Scholar] [CrossRef]
- Chang, J.; Shi, Y.; Si, G.; Yang, Q.; Dong, J.; Chen, J. The bioremediation potentials and mercury(II)-resistant mechanisms of a novel fungus Penicillium spp. DC-F11 isolated from contaminated soil. J. Hazard. Mater. 2020, 396, 122638. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.R.; Demarche, P.; Agathos, S.N. Formulation and characterization of an immobilized laccase biocatalyst and its application to eliminate organic micropollutants in wastewater. New Biotechnol. 2013, 30, 814–823. [Google Scholar] [CrossRef]
- Talukdar, D.; Jasrotia, T.; Sharma, R.; Jaglan, S.; Kumar, R.; Vats, R.; Kumar, R.; Mahnashi, M.H.; Umar, A. Evaluation of novel indigenous fungal consortium for enhanced bioremediation of heavy metals from contaminated sites. Environ. Technol. Innov. 2020, 20, 101050. [Google Scholar] [CrossRef]
- Garcia, L.; Lacerda, M.; Thomaz, D.; de Souza Golveia, J.; Pereira, M.; de Souza Gil, E.; Schimidt, F.; Santiago, M. laccase-alginate-chitosan-based matrix toward 17 α-ethinylestradiol removal. Prep. Biochem. Biotechnol. 2019, 49, 375–383. [Google Scholar] [CrossRef]
- Zhang, M.; Puri, A.K.; Govender, A.; Wang, Z.; Singh, S.; Permaul, K. The multi-chitinolytic enzyme system of the compost-dwelling thermophilic fungus Thermomyces lanuginosus. Process. Biochem. 2015, 50, 237–244. [Google Scholar] [CrossRef]
- Sosa-Martínez, J.D.; Balagurusamy, N.; Montañez, J.; Peralta, R.A.; Moreira, R.D.F.P.M.; Bracht, A.; Peralta, R.M.; Morales-Oyervides, L. Synthetic dyes biodegradation by fungal ligninolytic enzymes: Process optimization, metabolites evaluation and toxicity assessment. J. Hazard. Mater. 2020, 400, 123254. [Google Scholar] [CrossRef] [PubMed]
- Rajhans, G.; Sen, S.K.; Barik, A.; Raut, S. Elucidation of fungal dye-decolourizing peroxidase (DyP) and ligninolytic enzyme activities in decolourization and mineralization of azo dyes. J. Appl. Microbiol. 2020, 129, 1633–1643. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Zhou, G.; Peng, C.; Zhang, Y.; Kües, U.; Liu, J.; Xiao, Y.; Fang, Z. The first fungal laccase with an alkaline pH optimum obtained by directed evolution and its application in indigo dye decolorization. AMB Express 2019, 9, 151. [Google Scholar] [CrossRef]
- Li, H.-X.; Zhang, R.-J.; Tang, L.; Zhang, J.-H.; Mao, Z.-G. In vivo and in vitro decolorization of synthetic dyes by laccase from solid state fermentation with Trametes sp. SYBC-L4. Bioprocess Biosyst. Eng. 2014, 37, 2597–2605. [Google Scholar] [CrossRef]
- Emami, E.; Zolfaghari, P.; Golizadeh, M.; Karimi, A.; Lau, A.; Ghiasi, B.; Ansari, Z. Effects of stabilizers on sustainability, activity and decolorization performance of Manganese Peroxidase enzyme produced by Phanerochaete chrysosporium. J. Environ. Chem. Eng. 2020, 8, 104459. [Google Scholar] [CrossRef]
- Liu, S.; Ahmed, S.; Zhang, C.; Liu, T.; Shao, C.; Fang, Y. Diversity and antimicrobial activity of culturable fungi associated with sea anemone Anthopleura xanthogrammica. Electron. J. Biotechnol. 2020, 44, 41–46. [Google Scholar] [CrossRef]
- Zanutto-Elgui, M.R.; Vieira, J.C.S.; do Prado, D.Z.; Buzalaf, M.A.R.; de Magalhães Padilha, P.; de Oliveira, D.E.; Fleuri, L.F. Production of milk peptides with antimicrobial and antioxidant properties through fungal proteases. Food Chem. 2018, 278, 823–831. [Google Scholar] [CrossRef]
- Rafael, O.-H.D.; Fernándo, Z.-G.L.; Abraham, P.-T.; Alberto, V.-L.P.; Guadalupe, G.-S.; Pablo, P.J. Production of chitosan-oligosaccharides by the chitin-hydrolytic system of Trichoderma harzianum and their antimicrobial and anticancer effects. Carbohydr. Res. 2019, 486, 107836. [Google Scholar] [CrossRef] [PubMed]
- Abu-Tahon, M.A.; Isaac, G.S. Anticancer and antifungal efficiencies of purified chitinase produced from Trichoderma viride under submerged fermentation. J. Gen. Appl. Microbiol. 2020, 66, 32–40. [Google Scholar] [CrossRef] [Green Version]
- Baskar, G.; Chandhuru, J.; Fahad, K.S.; Praveen, A.S.; Chamundeeswari, M.; Muthukumar, T. Anticancer activity of fungal l-asparaginase conjugated with zinc oxide nanoparticles. J. Mater. Sci. Mater. Med. 2015, 26, 43. [Google Scholar] [CrossRef]
- Zhu, Z.; Yang, M.; Bai, Y.; Ge, F.; Wang, S. Antioxidant-related catalaseCTA1regulates development, aflatoxin biosynthesis, and virulence in pathogenic fungusAspergillus flavus. Environ. Microbiol. 2020, 22, 2792–2810. [Google Scholar] [CrossRef]
- El-Katony, T.M.; El-Dein, M.M.N.; El-Fallal, A.A.; Ibrahim, N.G.; Mousa, M. Substrate–fungus interaction on the enzymatic and non-enzymatic antioxidant activities of solid state fermentation system. Bioresour. Bioprocess. 2020, 7, 28. [Google Scholar] [CrossRef]
- Maitan-Alfenas, G.P.; Visser, E.M.; Guimarães, V.M. Enzymatic hydrolysis of lignocellulosic biomass: Converting food waste in valuable products. Curr. Opin. Food Sci. 2015, 1, 44–49. [Google Scholar] [CrossRef]
- Gupta, V.K.; Kubicek, C.P.; Berrin, J.-G.; Wilson, D.W.; Couturier, M.; Berlin, A.; Filho, E.X.; Ezeji, T. Fungal Enzymes for Bio-Products from Sustainable and Waste Biomass. Trends Biochem. Sci. 2016, 41, 633–645. [Google Scholar] [CrossRef] [Green Version]
- Jeske, L.; Placzek, S.; Schomburg, I.; Chang, A.; Schomburg, D. BRENDA in 2019: A European ELIXIR core data resource. Nucleic Acids Res. 2019, 47, D542–D549. [Google Scholar] [CrossRef]
- Berbee, M.L.; James, T.Y.; Strullu-Derrien, C. Early Diverging Fungi: Diversity and Impact at the Dawn of Terrestrial Life. Annu. Rev. Microbiol. 2017, 71, 41–60. [Google Scholar] [CrossRef] [Green Version]
- Verma, M.; Thakur, M.; Randhawa, J.; Sharma, D.; Thakur, A.; Meehnian, H.; Jana, A. Biotechnological applications of fungal enzymes with special reference to bioremediation. In Environmental Biotechnology; Gothandam, K.M., Ranjan, S., Lichtfouse, E., Dasgupta, N., Eds.; Springer: Cham, Switzerland, 2020; pp. 221–247. ISBN 9783030381967. [Google Scholar]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A Broader View: Microbial Enzymes and Their Relevance in Industries, Medicine, and Beyond. BioMed Res. Int. 2013, 2013, 329121. [Google Scholar] [CrossRef] [Green Version]
- Datta, P.; Gest, H. Control of enzyme activity by concerted feedback inhibition. Proc. Natl. Acad. Sci. USA 1964, 52, 1004–1009. [Google Scholar] [CrossRef] [Green Version]
- Lemieux, R.; Spohr, U. How Emil Fischer was led to the lock and key concept for enzyme specificity. Adv. Carbohydr. Chem. Biochem. 1994, 50, 1–20. [Google Scholar]
- Bhatia, S. Introduction to enzymes and their applications. In Introduction to Pharmaceutical Biotechnology: Enzymes, Proteins and Bioinformatics; Bhatia, S., Ed.; IOP Publishing: Bristol, UK, 2018; pp. 1–29. ISBN 9780750316026. [Google Scholar]
- Koshland, D.E. Das Schüssel-Schloß-Prinzip und die Induced-fit-Theorie. Angew. Chem. 1994, 106, 2468–2472. [Google Scholar] [CrossRef]
- Johnson, K.A.; Goody, R.S. The Original Michaelis Constant: Translation of the 1913 Michaelis–Menten Paper. Biochemistry 2011, 50, 8264–8269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyce, S.; Tipton, K.F. Enzyme Classification and Nomenclature. Encycl. Life Sci. 2013, 1, 1–11. [Google Scholar]
- arrett, A. Enzyme Nomenclature. Recommendations 1992. Eur. J. Biochem. 1995, 232, 1–6. [Google Scholar] [CrossRef]
- Yu, Q.-K.; Han, L.-T.; Wu, Y.-J.; Liu, T.-B. The Role of Oxidoreductase-Like Protein Olp1 in Sexual Reproduction and Virulence of Cryptococcus neoformans. Microorganisms 2020, 8, 1730. [Google Scholar] [CrossRef]
- Pedrini, N.; Ortiz-Urquiza, A.; Huarte-Bonnet, C.; Fan, Y.; Juárez, M.P.; Keyhani, N.O. Tenebrionid secretions and a fungal benzoquinone oxidoreductase form competing components of an arms race between a host and pathogen. Proc. Natl. Acad. Sci. USA 2015, 112, E3651–E3660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Góralczyk-Bińkowska, A.; Jasińska, A.; Długoński, A.; Płociński, P.; Długoński, J. Laccase activity of the ascomycete fungus Nectriella pironii and innovative strategies for its production on leaf litter of an urban park. PLoS ONE 2020, 15, e0231453. [Google Scholar] [CrossRef] [Green Version]
- Sayyed, R.Z.; Bhamare, H.M.; Sapna; Marraiki, N.; Elgorban, A.M.; Syed, A.; El-Enshasy, H.A.; Dailin, D.J. Tree bark scrape fungus: A potential source of laccase for application in bioremediation of non-textile dyes. PLOS ONE 2020, 15, e0229968. [Google Scholar] [CrossRef]
- Sharma, D.; Chaudhary, R.; Kaur, J.; Arya, S.K. Greener approach for pulp and paper industry by Xylanase and Laccase. Biocatal. Agric. Biotechnol. 2020, 25, 101604. [Google Scholar] [CrossRef]
- Shin, S.K.; Hyeon, J.E.; Joo, Y.-C.; Jeong, D.W.; You, S.K.; Han, S.O. Effective melanin degradation by a synergistic laccase-peroxidase enzyme complex for skin whitening and other practical applications. Int. J. Biol. Macromol. 2019, 129, 181–186. [Google Scholar] [CrossRef]
- Sorrentino, I.; Giardina, P.; Piscitelli, A. Development of a biosensing platform based on a laccase-hydrophobin chimera. Appl. Microbiol. Biotechnol. 2019, 103, 3061–3071. [Google Scholar] [CrossRef]
- Mehandia, S.; Sharma, S.C.; Arya, S.K. Isolation and characterization of an alkali and thermostable laccase from a novel Alcaligenes faecalis and its application in decolorization of synthetic dyes. Biotechnol. Rep. 2020, 25, e00413. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Fernández, M.; Sanromán, M. Ángeles; Moldes, D. Recent developments and applications of immobilized laccase. Biotechnol. Adv. 2013, 31, 1808–1825. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chang, F.; Tang, X.; Li, W.; Yin, Q.; Yang, Y.; Hu, Y. Bacterial laccase of Anoxybacillus ayderensis SK3-4 from hot springs showing potential for industrial dye decolorization. Ann. Microbiol. 2020, 70, 51. [Google Scholar] [CrossRef]
- Periasamy, D.; Mani, S.; Ambikapathi, R. White Rot Fungi and Their Enzymes for the Treatment of Industrial Dye Effluents. In Recent Advancement in White Biotechnology through Fungi; Yadav, A., Singh, S., Mishra, S., Gupta, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 73–100. ISBN 9783030255060. [Google Scholar]
- Manavalan, T.; Manavalan, A.; Heese, K. Characterization of Lignocellulolytic Enzymes from White-Rot Fungi. Curr. Microbiol. 2015, 70, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Erden, E.; Ucar, M.C.; Gezer, T.; Pazarlioglu, N.K. Screening for ligninolytic enzymes from autochthonous fungi and applications for decolorization of Remazole Marine Blue. Braz. J. Microbiol. 2009, 40, 346–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manubolu, M.; Goodla, L.; Pathakoti, K.; Malmlöf, K. Enzymes as direct decontaminating agents-mycotoxins. Enzym. Hum. Anim. Nutr. Princ. Perspect. 2018, 313–330. [Google Scholar] [CrossRef]
- Mehta, P.; Hale, T.I.; Christen, P. Aminotransferases: Demonstration of homology and division into evolutionary subgroups. JBIC J. Biol. Inorg. Chem. 1993, 214, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, Y.; Prosper, P.; Favier, F.; Harvengt, L.; Didierjean, C.; Jacquot, J.-P.; Morel-Rouhier, M.; Gelhaye, E. Diversification of Fungal Specific Class A Glutathione Transferases in Saprotrophic Fungi. PLOS ONE 2013, 8, e80298. [Google Scholar] [CrossRef]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef] [PubMed]
- Rahmanpour, S.; Backhouse, D.; Nonhebel, H. Induced tolerance of Sclerotinia sclerotiorum to isothiocyanates and toxic volatiles from Brassica species. Plant Pathol. 2009, 58, 479–486. [Google Scholar] [CrossRef]
- Calmes, B.; Morel-Rouhier, M.; Bataillé-Simoneau, N.; Gelhaye, E.; Guillemette, T.; Simoneau, P. Characterization of glutathione transferases involved in the pathogenicity of Alternaria brassicicola. BMC Microbiol. 2015, 15, 123. [Google Scholar] [CrossRef] [Green Version]
- Al-Madboly, L.; Ali, S.M.; El Fakharany, E.M.; Ragab, A.E.; Khedr, E.G.; Elokely, K.M. Stress-Based Production, and Characterization of Glutathione Peroxidase and Glutathione S-Transferase Enzymes from Lactobacillus plantarum. Front. Bioeng. Biotechnol. 2020, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Morel, M.; Ngadin, A.A.; Droux, M.; Jacquot, J.-P.; Gelhaye, E. The fungal glutathione S-transferase system. Evidence of new classes in the wood-degrading basidiomycete Phanerochaete chrysosporium. Cell. Mol. Life Sci. 2009, 66, 3711–3725. [Google Scholar] [CrossRef] [PubMed]
- Garcerá, A.; Barreto, L.; Piedrafita, L.; Tamarit, J.; Herrero, E. Saccharomyces cerevisiae cells have three Omega class glutathione S-transferases acting as 1-Cys thiol transferases. Biochem. J. 2006, 398, 187–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lateef, A.; Oloke, J.; Gueguim-Kana, E.; Raimi, O. Production of fructosyltransferase by a local isolate ofAspergillus nigerin both submerged and solid substrate media. Acta Aliment. 2012, 41, 100–117. [Google Scholar] [CrossRef]
- Nakakuki, T. Development of Functional Oligosaccharides in Japan. Trends Glycosci. Glycotechnol. 2003, 15, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Cunha, J.T.; Soares, P.O.; Romaní, A.; Thevelein, J.M.; Domingues, L. Biotechnology for Biofuels Xylose fermentation efficiency of industrial Saccharomyces cerevisiae yeast with separate or combined xylose reductase/xylitol dehydrogenase and xylose isomerase pathways. Biotechnol. Biofuels 2019, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Liu, Y.; Yang, J.; Ma, X.; Zeng, F.; Zhang, Z.; Wang, S.; Han, H.; Qin, H.; Lu, F. Cloning, expression and characterization of a novel fructosyltransferase from Aspergillus niger and its application in the synthesis of fructooligosaccharides. RSC Adv. 2019, 9, 23856–23863. [Google Scholar] [CrossRef] [Green Version]
- Ghazi, I.; Fernandez-Arrojo, L.; Garcia-Arellano, H.; Ferrer, M.; Ballesteros, A.O.; Plou, F.J. Purification and kinetic characterization of a fructosyltransferase from Aspergillus aculeatus. J. Biotechnol. 2007, 128, 204–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saranraj, P.; Stella, D. Fungal amylase—A review. Int. J. Microbiol. Res. 2013, 4, 203–211. [Google Scholar] [CrossRef]
- Singh, S. Aspergillus Enzymes for Textile Industry. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier BV: Amsterdam, The Netherlands, 2016; pp. 191–198. [Google Scholar]
- Chimbekujwo, K.I.; Ja’Afaru, M.I.; Adeyemo, O.M. Purification, characterization and optimization conditions of protease produced by Aspergillus brasiliensis strain BCW. Sci. Afr. 2020, 8, e00398. [Google Scholar] [CrossRef]
- Yike, I. Fungal Proteases and Their Pathophysiological Effects. Mycopathologia 2011, 171, 299–323. [Google Scholar] [CrossRef] [PubMed]
- Germano, S.; Pandey, A.; Osaku, C.A.; Rocha, S.N.; Soccol, C.R. Characterization and stability of proteases from Penicillium sp. produced by solid-state fermentation. Enzym. Microb. Technol. 2003, 32, 246–251. [Google Scholar] [CrossRef]
- Vishwanatha, K.S.; Rao, A.G.A.; Singh, S.A. Acid protease production by solid-state fermentation using Aspergillus oryzae MTCC 5341: Optimization of process parameters. J. Ind. Microbiol. Biotechnol. 2010, 37, 129–138. [Google Scholar] [CrossRef] [PubMed]
- El-Shora, H.M.; Metwally, M.A.-A. Production, purification and characterisation of proteases from whey by some fungi. Ann. Microbiol. 2008, 58, 495–502. [Google Scholar] [CrossRef]
- De Souza, P.M.; Bittencourt, M.L.D.A.; Caprara, C.C.; de Freitas, M.; de Almeida, R.P.C.; Silveira, D.; Fonseca, Y.M.; Ferreira Filho, E.X.; Pessoa Junior, A.; Magalhães, P.O. A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 2015, 46, 337–346. [Google Scholar] [CrossRef] [Green Version]
- Benluvankar, V.; Jebapriya, G.R.; Gnanadoss, J.J. Protease production by Penicillium sp. LCJ228 under solid state fermentation using groundnut oilcake as substrate. Int. J. Life Sci. Pharma Res. 2015, 5, 12–19. [Google Scholar]
- Pekkarinen, A.; Mannonen, L.; Jones, B.; Niku-Paavola, M.-L. Production of Proteases by Fusarium Species Grown on Barley Grains and in Media Containing Cereal Proteins. J. Cereal Sci. 2000, 31, 253–261. [Google Scholar] [CrossRef]
- Al-Askar, A.; Abdulkhair, W.; Rashad, Y. Production, Purification and Production, Purification and Optimization of Protease by Fusarium solani under Solid State Fermentation and Isolation of Protease Inhibitor Protein from Rumex vesicarius L. J. Pure Appl. Microbiol. 2014, 8, 239–250. [Google Scholar]
- Kim, J.Y. Isolation of Protease-producing Yeast, Pichia farinosa CO-2 and Characterization of Its Extracellular Enzyme. J. Korean Soc. Appl. Biol. Chem. 2010, 53, 133–141. [Google Scholar] [CrossRef]
- Zakowski, J.J.; Bruns, D.E. Biochemistry of Human Alpha Amylase Isoenzymes. CRC Crit. Rev. Clin. Lab. Sci. 1985, 21, 283–322. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.; Bhatti, H.N.; Pervin, F. Production, partial purification and thermal characterization of β-Amylase from Fusarium solani in solid state fermentation. J. Chem. Soc. Pakistan 2008, 30, 480–485. [Google Scholar]
- Oseni, O.A. Activity of β-Amylase in Some Fungi Strains Isolated from Forest Soil in South-Western Nigeria. Br. Biotechnol. J. 2014, 4, 1–9. [Google Scholar] [CrossRef]
- Silano, V.; Barat Baviera, J.M.; Bolognesi, C.; Cocconcelli, P.S.; Crebelli, R.; Gott, D.M.; Grob, K.; Lampi, E.; Mortensen, A.; Rivière, G.; et al. Safety evaluation of the food enzyme α-amylase from Aspergillus oryzae (strain DP-Bzb41). EFSA J. 2019, 17, e05899. [Google Scholar] [CrossRef]
- Ahmed, N.E.; El Shamy, A.R.; Awad, H.M. Optimization and immobilization of amylase produced by Aspergillus terreus using pomegranate peel waste. Bull. Natl. Res. Cent. 2020, 44, 109. [Google Scholar] [CrossRef]
- Bakri, Y.; Jawhar, M.; Arabi, M.I.E. Enhanced amylase production by Fusarium solani in solid state fermentation. Pak. J. Sci. Ind. Res. Ser. B Biol. Sci. 2014, 57, 123–128. [Google Scholar] [CrossRef]
- Shruthi, B.R.; Achur, R.N.H.; Nayaka Boramuthi, T. Optimized Solid-State Fermentation Medium Enhances the Multienzymes Production from Penicillium citrinum and Aspergillus clavatus. Curr. Microbiol. 2020, 77, 2192–2206. [Google Scholar] [CrossRef] [PubMed]
- Kanmani, P.; Aravind, J.; Kumaresan, K. An insight into microbial lipases and their environmental facet. Int. J. Environ. Sci. Technol. 2014, 12, 1147–1162. [Google Scholar] [CrossRef] [Green Version]
- Bora, L. Purification and Characterization of Highly Alkaline Lipase from Bacillus licheniformis MTCC 2465: And Study of its Detergent Compatibility and Applicability. J. Surfactants Deterg. 2014, 17, 889–898. [Google Scholar] [CrossRef]
- Abdel-Fattah, Y.; Soliman, N.A.; Yousef, S.M.; El-Helow, E.R. Application of experimental designs to optimize medium composition for production of thermostable lipase/esterase by Geobacillus thermodenitrificans AZ1. J. Genet. Eng. Biotechnol. 2012, 10, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Ilesanmi, O.I.; Adekunle, A.E.; Omolaiye, J.A.; Olorode, E.M.; Ogunkanmi, A.L. Isolation, optimization and molecular characterization of lipase producing bacteria from contaminated soil. Sci. Afr. 2020, 8, e00279. [Google Scholar] [CrossRef]
- Nema, A.; Patnala, S.H.; Mandari, V.; Kota, S.; Devarai, S.K. Production and optimization of lipase using Aspergillus niger MTCC 872 by solid-state fermentation. Bull. Natl. Res. Cent. 2019, 43, 82. [Google Scholar] [CrossRef] [Green Version]
- Putri, D.N.; Khootama, A.; Perdani, M.S.; Utami, T.S.; Hermansyah, H. Optimization of Aspergillus niger lipase production by solid state fermentation of agro-industrial waste. Energy Rep. 2020, 6, 331–335. [Google Scholar] [CrossRef]
- Kempka, A.P.; Lipke, N.L.; Da Luz Fontoura Pinheiro, T.; Menoncin, S.; Treichel, H.; Freire, D.M.G.; Di Luccio, M.; De Oliveira, D. Response surface method to optimize the production and characterization of lipase from Penicillium verrucosum in solid-state fermentation. Bioprocess Biosyst. Eng. 2008, 31, 119–125. [Google Scholar] [CrossRef]
- Geoffry, K.; Achur, R.N. Optimization of novel halophilic lipase production by Fusarium solani strain NFCCL 4084 using palm oil mill effluent. J. Genet. Eng. Biotechnol. 2018, 16, 327–334. [Google Scholar] [CrossRef]
- El Aamri, L.; Hafidi, M.; Scordino, F.; Krasowska, A.; Lebrihi, A.; Orlando, M.G.; Barresi, C.; Criseo, G.; Barreca, D.; Romeo, O. Arthrographis curvata and Rhodosporidium babjevae as New Potential Fungal Lipase Producers for Biotechnological Applications. Braz. Arch. Biol. Technol. 2020, 63, 1–9. [Google Scholar] [CrossRef]
- Salihu, A.; Bala, M.; Bala, S.M. Application of Plackett-Burman Experimental Design for Lipase Production by Aspergillus niger Using Shea Butter Cake. ISRN Biotechnol. 2013, 2013, 718352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleuri, L.F.; de Oliveira, M.C.; de Lara Campos Arcuri, M.; Capoville, B.L.; Pereira, M.S.; Delgado, C.H.O.; Novelli, P.K. Production of fungal lipases using wheat bran and soybean bran and incorporation of sugarcane bagasse as a co-substrate in solid-state fermentation. Food Sci. Biotechnol. 2014, 23, 1199–1205. [Google Scholar] [CrossRef]
- Mehta, A.; Bodh, U.; Gupta, R. Fungal lipases: A review. J. Biotech Res. 2017, 8, 58–77. [Google Scholar]
- Selvakumar, P.; Sivashanmugam, P. Optimization of lipase production from organic solid waste by anaerobic digestion and its application in biodiesel production. Fuel Process. Technol. 2017, 165, 1–8. [Google Scholar] [CrossRef]
- Gopinath, S.C.B.; Anbu, P.; Lakshmipriya, T.; Hilda, A. Strategies to Characterize Fungal Lipases for Applications in Medicine and Dairy Industry. BioMed Res. Int. 2013, 2013, 154549. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Fact. 2020, 19, 169. [Google Scholar] [CrossRef]
- Hwang, H.T.; Qi, F.; Yuan, C.; Zhao, X.; Ramkrishna, D.; Liu, D.; Varma, A. Lipase-catalyzed process for biodiesel production: Protein engineering and lipase production. Biotechnol. Bioeng. 2014, 111, 639–653. [Google Scholar] [CrossRef]
- Meyer, V.; Andersen, M.R.; Brakhage, A.A.; Braus, G.H.; Caddick, M.X.; Cairns, T.C.; De Vries, R.P.; Haarmann, T.; Hansen, K.; Hertz-Fowler, C.; et al. Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: A white paper. Fungal Biol. Biotechnol. 2016, 3, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Várnai, A.; Mäkelä, M.R.; Djajadi, D.T.; Rahikainen, J.; Hatakka, A.; Viikari, L. Carbohydrate-binding modules of fungal cellulases: Occurrence in nature, function, and relevance in industrial biomass conversion. Adv. Appl. Microbiol. 2014, 88, 103–165. [Google Scholar]
- Abdullah, A.; Hamid, H.; Christwardana, M.; Hadiyanto, H. Optimization of Cellulase Production by Aspergillus niger ITBCC L74 with Bagasse as Substrate using Response Surface Methodology. HAYATI J. Biosci. 2018, 25, 115. [Google Scholar] [CrossRef]
- Amadi, O.C.; Egong, E.J.; Nwagu, T.N.; Okpala, G.; Onwosi, C.O.; Chukwu, G.C.; Okolo, B.N.; Agu, R.C.; Moneke, A.N. Process optimization for simultaneous production of cellulase, xylanase and ligninase by Saccharomyces cerevisiae SCPW 17 under solid state fermentation using Box-Behnken experimental design. Heliyon 2020, 6, e04566. [Google Scholar] [CrossRef] [PubMed]
- Cekmecelioglu, D.; Demirci, A. Production of Cellulase and Xylanase Enzymes Using Distillers Dried Grains with Solubles (DDGS) by Trichoderma reesei at Shake-Flask Scale and the Validation in the Benchtop Scale Bioreactor. Waste Biomass-Valorization 2020, 11, 6575–6584. [Google Scholar] [CrossRef]
- Polizeli, M.L.T.M.; Rizzatti, A.C.S.; Monti, R.; Terenzi, H.F.; Jorge, J.A.; Amorim, D.S. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005, 67, 577–591. [Google Scholar] [CrossRef]
- Silva, L.; Terrasan, C.R.F.; Carmona, E.C. Purification and characterization of xylanases from Trichoderma inhamatum. Electron. J. Biotechnol. 2015, 18, 307–313. [Google Scholar] [CrossRef] [Green Version]
- Khalaji, A.; Sedighi, M.; Vahabzadeh, F. Optimization and Kinetic Evaluation of Acetylxylan Esterase and Xylanase Production by Trichoderma reesei Using Corn Cob Xylan. Environ. Process. 2020, 7, 885–909. [Google Scholar] [CrossRef]
- Atalla, S.M.M.; Ahmed, N.E.; Awad, H.M.; El Gamal, N.G.; El Shamy, A.R. Statistical optimization of xylanase production, using different agricultural wastes by Aspergillus oryzae MN894021, as a biological control of faba bean root diseases. Egypt. J. Biol. Pest Control. 2020, 30, 125. [Google Scholar] [CrossRef]
- Nikhil, B.; Dharmesh, A.; Priti, T. International Journal of Research in Chemistry and Environment Production of Xylanase by Aspergillus flavus FPDN1 on Pearl millet bran: Optimization of Culture Conditions and Application in Bioethanol Production. Int. J. Res. Chem. Environ. 2012, 2, 204–210. [Google Scholar]
- Corrêa, T.L.R.; de Araújo, E.F. Fungal phytases: From genes to applications. Braz. J. Microbiol. 2020, 51, 1009–1020. [Google Scholar] [CrossRef]
- Kc, S.; Upadhyaya, J.; Joshi, D.R.; Lekhak, B.; Chaudhary, D.K.; Pant, B.R.; Bajgai, T.R.; Dhital, R.; Khanal, S.; Koirala, N.; et al. Production, Characterization, and Industrial Application of Pectinase Enzyme Isolated from Fungal Strains. Fermentation 2020, 6, 59. [Google Scholar] [CrossRef]
- Mathew, G.M.; Madhavan, A.; Arun, K.B.; Sindhu, R.; Binod, P.; Singhania, R.R.; Sukumaran, R.K.; Pandey, A. Thermophilic Chitinases: Structural, Functional and Engineering Attributes for Industrial Applications. Appl. Biochem. Biotechnol. 2021, 193, 142–164. [Google Scholar] [CrossRef] [PubMed]
- Hartl, L.; Zach, S.; Seidl-Seiboth, V. Fungal chitinases: Diversity, mechanistic properties and biotechnological potential. Appl. Microbiol. Biotechnol. 2011, 93, 533–543. [Google Scholar] [CrossRef] [Green Version]
- Elsoud, M.M.A.; El Kady, E.M. Current trends in fungal biosynthesis of chitin and chitosan. Bull. Natl. Res. Cent. 2019, 43, 59. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.-H.; Fu, X.; Yan, X.-Y.; Peng, W.-F.; Kang, L.-X. A Broad-Specificity Chitinase from Penicillium oxalicum k10 Exhibits Antifungal Activity and Biodegradation Properties of Chitin. Mar. Drugs 2021, 19, 356. [Google Scholar] [CrossRef] [PubMed]
- Omumasaba, C.A.; Yoshida, N.; Ogawa, K. Purification and characterization of a chitinase from Trichoderma viride. J. Gen. Appl. Microbiol. 2001, 47, 53–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krolicka, M.; Hinz, S.W.A.; Koetsier, M.J.; Joosten, R.; Eggink, G.; Van Den Broek, L.A.M.; Boeriu, C.G. Chitinase Chi1 from Myceliophthora thermophila C1, a Thermostable Enzyme for Chitin and Chitosan Depolymerization. J. Agric. Food Chem. 2018, 66, 1658–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.; Brar, A.; Vivekanand, V.; Pareek, N. Production of chitinase from thermophilic Humicola grisea and its application in production of bioactive chitooligosaccharides. Int. J. Biol. Macromol. 2017, 104, 1641–1647. [Google Scholar] [CrossRef]
- Baldoni, D.B.; Antoniolli, Z.I.; Mazutti, M.A.; Jacques, R.J.S.; Dotto, A.C.; de Oliveira Silveira, A.; Ferraz, R.C.; Soares, V.B.; de Souza, A.R.C. Chitinase production by Trichoderma koningiopsis UFSMQ40 using solid state fermentation. Braz. J. Microbiol. 2020, 51, 1897–1908. [Google Scholar] [CrossRef] [PubMed]
- Sathiyaraj, G.; Srinivasan, S.; Kim, H.-B.; Subramaniyam, S.; Lee, O.R.; Kim, Y.-J.; Yang, D.C. Screening and optimization of pectin lyase and polygalacturonase activity from ginseng pathogen Cylindrocarpon Destructans. Braz. J. Microbiol. 2011, 42, 794–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atanasova, L.; Dubey, M.; Grujić, M.; Gudmundsson, M.; Lorenz, C.; Sandgren, M.; Kubicek, C.P.; Jensen, D.F.; Karlsson, M. Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family. BMC Microbiol. 2018, 18, 178. [Google Scholar] [CrossRef]
- Semenova, M.V.; Sinitsyna, O.A.; Morozova, V.V.; Fedorova, E.A.; Gusakov, A.V.; Okunev, O.N.; Sokolova, L.M.; Koshelev, A.V.; Bubnova, T.V.; Vinetskii, Y.P.; et al. Use of a preparation from fungal pectin lyase in the food industry. Appl. Biochem. Microbiol. 2006, 42, 598–602. [Google Scholar] [CrossRef]
- Singh, R.P.; Gupta, V.; Kumari, P.; Kumar, M.; Reddy, C.R.K.; Prasad, K.; Jha, B. Purification and partial characterization of an extracellular alginate lyase from Aspergillus oryzae isolated from brown seaweed. Environ. Boil. Fishes 2010, 23, 755–762. [Google Scholar] [CrossRef]
- Pilgaard, B.; Wilkens, C.; Herbst, F.-A.; Vuillemin, M.; Rhein-Knudsen, N.; Meyer, A.S.; Lange, L. Proteomic enzyme analysis of the marine fungus Paradendryphiella salina reveals alginate lyase as a minimal adaptation strategy for brown algae degradation. Sci. Rep. 2019, 9, 12338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nwokoro, O. Studies on the production of glucose isomerase by Bacillus licheniformis. Pol. J. Chem. Technol. 2015, 17, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Crueger, A.; Crueger, W. Glucose transforming enzymes. In Microbial Enzymes and Biotechnology; Fogarty, W.M., Kelly, C.T., Eds.; Springer: Dordrecht, The Netherlands, 1990; pp. 177–226. ISBN 978-94-009-0765-2. [Google Scholar]
- Bhosale, S.H.; Rao, M.B.; Deshpande, V.V. Molecular and industrial aspects of glucose isomerase. Microbiol. Rev. 1996, 60, 280–300. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.K.; Xu, C.; Qin, W. Isolation of Bacterial Strain with Xylanase and Xylose/Glucose Isomerase (GI) Activity and Whole Cell Immobilization for Improved Enzyme Production. Waste Biomass-Valorization 2021, 12, 833–845. [Google Scholar] [CrossRef]
- Dai, C.; Miao, T.; Hai, J.; Xiao, Y.; Li, Y.; Zhao, J.; Qiu, H.; Xu, B. A Novel Glucose Isomerase fromCaldicellulosiruptor besciiwith Great Potentials in the Production of High-Fructose Corn Syrup. BioMed Res. Int. 2020, 2020, 1871934. [Google Scholar] [CrossRef] [Green Version]
- Rengasamy, S.; Subramanian, M.R.; Perumal, V.; Ganeshan, S.; Al Khulaifi, M.M.; Al-Shwaiman, H.A.; Elgorban, A.M.; Syed, A.; Thangaprakasam, U. Purification and kinetic behavior of glucose isomerase from Streptomyces lividans RSU. Saudi J. Biol. Sci. 2020, 27, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Sayyed, R.Z.; Shimpi, G.B.; Chincholkar, S.B. Constitutive production of extracellular glucose isomerase by an osmophillic Aspergillus sp. under submerged conditions. J. Food Sci. Technol. 2010, 47, 496–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, R.O.; Kooi, E.R. Enzymatic conversion of D-glucose to D-fructose. Science 1957, 125, 648–649. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.; Jin, Y.-S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: A review and perspective. Microb. Cell Factories 2017, 16, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lilius, S.T.A. Ethanolic Fermentation of Xylose with Saccharomyces cerevisiae Harboring the Thermus thermophilus xylA Gene, Which Expresses an Active Xylose (Glucose) Isomerase. Microb. Cell Factories 1996, 62, 4648–4651. [Google Scholar]
- Brat, D.; Boles, E.; Wiedemann, B. Functional Expression of a Bacterial Xylose Isomerase in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2009, 75, 2304–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidrauski, C.; Cox, J.S.; Walter, P. tRNA Ligase Is Required for Regulated mRNA Splicing in the Unfolded Protein Response. Cell 1996, 87, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, A.; Ghosh, S.; Goldgur, Y.; Shuman, S. Structure and two-metal mechanism of fungal tRNA ligase. Nucleic Acids Res. 2018, 47, 1428–1439. [Google Scholar] [CrossRef] [PubMed]
- Remus, B.S.; Schwer, B.; Shuman, S. Characterization of the tRNA ligases of pathogenic fungi Aspergillus fumigatus and Coccidioides immitis. RNA 2016, 22, 1500–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of Proteins by Ubiquitin and Ubiquitin-Like Proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulathu, Y.; Komander, D. Atypical ubiquitylation — the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Marín, I. Origin and evolution of fungal HECT ubiquitin ligases. Sci. Rep. 2018, 8, 6419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva, R.R. Enzyme technology in food preservation: A promising and sustainable strategy for biocontrol of post-harvest fungal pathogens. Food Chem. 2019, 277, 531–532. [Google Scholar] [CrossRef]
- Thomas, A.; Thomas, A. Acrylamide—A Potent Carcinogen in Food. Int. J. Sci. Res. 2014, 3, 177–188. [Google Scholar]
- Cladière, M.; Camel, V. The Maillard reaction and food safety: Focus on acrylamide. Environ. Risques St. 2017, 16, 31–43. [Google Scholar] [CrossRef]
- Swanston, J. Acrylamide Mitigation in Foods—An Update Acrylamide Mitigation in Foods—An Update; Leatherhead Food Research: Epsom, UK, 2019. [Google Scholar]
- Batool, T.; Makky, E.A.; Jalal, M.; Yusoff, M. A Comprehensive Review on l-Asparaginase and Its Applications. Appl. Biochem. Biotechnol. 2015, 178, 900–923. [Google Scholar] [CrossRef] [Green Version]
- Orabi, H.M.; El Fakharany, E.; Abdelkhalek, E.S.; Sidkey, N.M. l-Asparaginase And l-Glutaminase: Sources, Production, And Applications in Medicine and Industry. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 179–190. [Google Scholar] [CrossRef]
- Walia, A.; Guleria, S.; Mehta, P.; Chauhan, A.; Parkash, J. Microbial xylanases and their industrial application in pulp and paper biobleaching: A review. 3 Biotech 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.S.; Singh, T.; Pandey, A. Microbial enzymes–An overview. In Biomass, Biofuels, Biochemicals: Advances in Enzyme Technology; Elsevier: Amsterdam, The Netherlands, 2019; ISBN 9780444641144. [Google Scholar]
- Khonzue, P.; Laothanachareon, T.; Rattanaphan, N.; Tinnasulanon, P.; Apawasin, S.; Paemanee, A.; Ruanglek, V.; Tanapongpipat, S.; Champreda, V.; Eurwilaichitr, L. Optimization of Xylanase Production fromAspergillus nigerfor Biobleaching of Eucalyptus Pulp. Biosci. Biotechnol. Biochem. 2011, 75, 1129–1134. [Google Scholar] [CrossRef]
- Kumar, A.; Gautam, A.; Dutt, D. Bio-pulping: An energy saving and environment-friendly approach. Phys. Sci. Rev. 2020, 5. [Google Scholar] [CrossRef]
- Weng, C.; Peng, X.; Han, Y. Depolymerization and conversion of lignin to value-added bioproducts by microbial and enzymatic catalysis. Biotechnol. Biofuels 2021, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Maalej-Achouri, I.; Guerfali, M.; Romdhane, I.B.-B.; Gargouri, A.; Belghith, H. The effect of Talaromyces thermophilus cellulase-free xylanase and commercial laccase on lignocellulosic components during the bleaching of kraft pulp. Int. Biodeterior. Biodegrad. 2012, 75, 43–48. [Google Scholar] [CrossRef]
- Pensupa, N.; Leu, S.; Hu, Y.; Du, C.; Liu, H.; Jing, H.; Wang, H.; Lin, C. Recent Trends in Sustainable Textile Waste Recycling Methods: Current Situation and Future Prospects. In Chemistry and Chemical Technologies in Waste Valorization; Lin, C.S.K., Ed.; Springer: Cham, Switzerland, 2017; pp. 189–228. [Google Scholar]
- Araújo, R.; Casal, M.; Cavaco-Paulo, A. Application of enzymes for textile fibres processing. Biocatal. Biotransform. 2008, 26, 332–349. [Google Scholar] [CrossRef] [Green Version]
- Lopes, L.S.; Vieira, N.; da Luz, J.M.R.; Marliane de Cássia, S.S.; Cardoso, W.S.; Kasuya, M.C.M. Production of fungal enzymes in Macaúba coconut and enzymatic degradation of textile dye. Biocatal. Agric. Biotechnol. 2020, 26, 101651. [Google Scholar] [CrossRef]
- Velázquez-Fernández, J.B.; Muñiz-Hernández, S. Bioremediation: Processes, Challenges and Future Prospects; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2014; ISBN 9781629485157. [Google Scholar]
- Zhang, S.; Gedalanga, P.B.; Mahendra, S. Advances in bioremediation of 1,4-dioxane-contaminated waters. J. Environ. Manag. 2017, 204, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Santacruz-Juárez, E.; Buendia-Corona, R.E.; Ramírez, R.E.; Sánchez, C. Fungal enzymes for the degradation of polyethylene: Molecular docking simulation and biodegradation pathway proposal. J. Hazard. Mater. 2021, 411, 125118. [Google Scholar] [CrossRef]
- Steliga, T. Role of Fungi in Biodegradation of Petroleum Hydrocarbons. Pol. J. Environ. Stud. 2012, 21, 471–479. [Google Scholar]
- Sun, K.; Song, Y.; He, F.; Jing, M.; Tang, J.; Liu, R. A review of human and animals exposure to polycyclic aromatic hydrocarbons: Health risk and adverse effects, photo-induced toxicity and regulating effect of microplastics. Sci. Total Environ. 2021, 773, 145403. [Google Scholar] [CrossRef]
- Asemoloye, M.D.; Tosi, S.; Daccò, C.; Wang, X.; Xu, S.; Marchisio, M.A.; Gao, W.; Jonathan, S.G.; Pecoraro, L. Hydrocarbon Degradation and Enzyme Activities of Aspergillus oryzae and Mucor irregularis Isolated from Nigerian Crude Oil-Polluted Sites. Microorganisms 2020, 8, 1912. [Google Scholar] [CrossRef] [PubMed]
- Sanghi, R.; Dixit, A.; Verma, P.; Puri, S. Design of reaction conditions for the enhancement of microbial degradation of dyes in sequential cycles. J. Environ. Sci. 2009, 21, 1646–1651. [Google Scholar] [CrossRef]
- Rápó, E.; Tonk, S. Factors Affecting Synthetic Dye Adsorption; Desorption Studies: A Review of Results from the Last Five Years (2017–2021). Molecules 2021, 26, 5419. [Google Scholar] [CrossRef]
- Karnwal, A.; Singh, S.; Kumar, V.; Sidhu, G.; Dhanjal, D.; Datta, S.; Amin, D.; Saini, M.; Singh, J. Fungal enzymes for the textile industry. In Recent Advancement in White Biotechnology through Fungi; Yadav, A., Mishra, S., Singh, S., Gupta, A., Eds.; Springer: Cham, Switzerland, 2019; pp. 459–482. ISBN 978-3-030-10479-5. [Google Scholar]
- El Fakharany, E.; Hassan, M.A.; Taha, T.H. Production and Application of Extracellular Laccase Produced by Fusarium oxysporum EMT. Int. J. Agric. Biol. 2016, 18, 939–947. [Google Scholar] [CrossRef]
- Li, K.; Guan, G.; Zhu, J.; Wu, H.; Sun, Q. Antibacterial activity and mechanism of a laccase-catalyzed chitosan–gallic acid derivative against Escherichia coli and Staphylococcus aureus. Food Control 2019, 96, 234–243. [Google Scholar] [CrossRef]
- Fuglsang, C.C.; Johansen, C.; Christgau, S.; Adler-Nissen, J. Antimicrobial enzymes: Applications and future potential in the food industry. Trends Food Sci. Technol. 1995, 6, 390–396. [Google Scholar] [CrossRef]
- El Fakharany, E.; Haroun, B.M.; Ng, T.; Redwan, E. Oyster Mushroom Laccase Inhibits Hepatitis C Virus Entry into Peripheral Blood Cells and Hepatoma Cells. Protein Pept. Lett. 2010, 17, 1031–1039. [Google Scholar] [CrossRef]
- Li, H.-C.; Huang, E.-Y.; Su, P.-Y.; Wu, S.-Y.; Yang, C.-C.; Lin, Y.-S.; Chang, W.-C.; Shih, C. Nuclear Export and Import of Human Hepatitis B Virus Capsid Protein and Particles. PLOS Pathog. 2010, 6, e1001162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Zhao, L.; Wang, H.; Ng, T.B. A novel ribonuclease with antiproliferative activity toward leukemia and lymphoma cells and HIV-1 reverse transcriptase inhibitory activity from the mushroom, Hohenbuehelia serotina. Int. J. Mol. Med. 2014, 33, 209–214. [Google Scholar] [CrossRef] [Green Version]
- El-Maradny, Y.A.; El-Fakharany, E.M.; Abu-Serie, M.M.; Hashish, M.H.; Selim, H.S. Lectins purified from medicinal and edible mushrooms: Insights into their antiviral activity against pathogenic viruses. Int. J. Biol. Macromol. 2021, 179, 239–258. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Su, D.; Liu, Q.; Gao, W.; Kang, Y. Mushroom lectin overcomes hepatitis B virus tolerance via TLR6 signaling. Sci. Rep. 2017, 7, 5814. [Google Scholar] [CrossRef] [Green Version]
- Wang, H. Purification and characterization of a laccase from the edible wild mushroom Tricholoma mongolicum. J. Microbiol. Biotechnol. 2010, 20, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, L.G.; Matei, E.; LaSala, F.; Delgado, R.; Gronenborn, A.M. Dissecting carbohydrate–Cyanovirin-N binding by structure-guided mutagenesis: Functional implications for viral entry inhibition. Protein Eng. Des. Sel. 2006, 19, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-B.; Xu, B.-Y.; Huang, M.; Sun, L.-H.; Yang, Q.; Chen, Y.-J.; Yin, Y.-L.; He, Q.-G.; Sun, H. Adjuvant effects mediated by the carbohydrate recognition domain of Agrocybe aegerita lectin interacting with avian influenza H9N2 viral surface glycosylated proteins. J. Zhejiang Univ. Sci. B 2017, 18, 653–661. [Google Scholar] [CrossRef] [Green Version]
- Li, J.W.-H.; Vederas, J.C. Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef] [Green Version]
- Liaqat, F.; Eltem, R. Chitooligosaccharides and their biological activities: A comprehensive review. Carbohydr. Polym. 2018, 184, 243–259. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, H.; Barrero, R.A.; Zhang, B.; Sun, G.; Wilson, I.W.; Xie, F.; Walker, K.D.; Parks, J.W.; Bruce, R.; et al. Genome sequencing and analysis of the paclitaxel-producing endophytic fungus Penicillium aurantiogriseum NRRL. BMC Genom. 2014, 15, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, K.G.; Manikandan, R.; Arulvasu, C.; Pandi, M. Anti-proliferative effect of fungal taxol extracted from Cladosporium oxysporum against human pathogenic bacteria and human colon cancer cell line HCT 15. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 138, 667–674. [Google Scholar] [CrossRef]
- Zaiyou, J.; Li, M.; Xiqiao, H. An endophytic fungus efficiently producing paclitaxel isolated from Taxus wallichiana var. mairei. Medicine 2017, 96, e7406. [Google Scholar] [CrossRef]
- Ranjan, A.; Singh, R.K.; Singh, M. Metabolic versatility of fungi as a source for anticancer compounds. In Evolutionary Diversity as a Source for Anticancer Molecules; Srivastava, A.K., Kannaujiya, V.K., Singh, R.K., Singh, D., Eds.; Elsevier: London, UK, 2020; pp. 191–207. ISBN 9780128217108. [Google Scholar]
- Gallego-Jara, J.; Lozano-Terol, G.; Sola-Martínez, R.A.; Cánovas-Díaz, M.; de Diego Puente, T. A Compressive Review about Taxol®: History and Future Challenges. Molecules 2020, 25, 5986. [Google Scholar] [CrossRef]
- How, C.W.; Ong, Y.S.; Low, S.S.; Pandey, A.; Show, P.L.; Foo, J.B. How far have we explored fungi to fight cancer? Semin. Cancer Biol. 2021, in press. [CrossRef] [PubMed]
- Xu, L.; Liu, X.; Li, Y.; Yin, Z.; Jin, L.; Lu, L.; Qu, J.; Xiao, M. Enzymatic rhamnosylation of anticancer drugs by an α-l-rhamnosidase from Alternaria sp. L1 for cancer-targeting and enzyme-activated prodrug therapy. Appl. Microbiol. Biotechnol. 2019, 103, 7997–8008. [Google Scholar] [CrossRef] [PubMed]
- Stolworthy, T.S.; Korkegian, A.M.; Willmon, C.; Ardiani, A.; Cundiff, J.; Stoddard, B.L.; Black, M.E. Yeast Cytosine Deaminase Mutants with Increased Thermostability Impart Sensitivity to 5-Fluorocytosine. J. Mol. Biol. 2008, 377, 854–869. [Google Scholar] [CrossRef] [Green Version]
- Aklakur, M. Natural antioxidants from sea: A potential industrial perspective in aquafeed formulation. Rev. Aquac. 2016, 10, 385–399. [Google Scholar] [CrossRef]
- Butkhup, L.; Samappito, W.; Jorjong, S. Evaluation of bioactivities and phenolic contents of wild edible mushrooms from northeastern Thailand. Food Sci. Biotechnol. 2017, 27, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Rafi, S.; Shoaib, A.; Awan, Z.A.; Rizvi, N.B.; Nafisa; Shafiq, M. Chromium tolerance, oxidative stress response, morphological characteristics, and FTIR studies of phytopathogenic fungus Sclerotium rolfsii. Folia Microbiol. 2016, 62, 207–219. [Google Scholar] [CrossRef]
- Abdel-Azeem, M.; El-Maradny, Y.; Othman, A.; Abdel-Azeem, A. Endophytic Fungi as a Source of New Pharmaceutical Biomolecules. In Industrially Important Fungi for Sustainable Development. Fungal Biology; Abdel-Azeem, A.M., Yadav, A.N., Yadav, N., Sharma, M., Eds.; Springer: Cham, Switzerland, 2021; pp. 115–151. ISBN 9783030856038. [Google Scholar]
- Filipe, D.; Fernandes, H.; Castro, C.; Peres, H.; Oliva-Teles, A.; Belo, I.; Salgado, J.M. Improved lignocellulolytic enzyme production and antioxidant extraction using solid-state fermentation of olive pomace mixed with winery waste. Improv. Lignocellulolytic 2020, 14, 78–91. [Google Scholar] [CrossRef] [Green Version]
- Arnau, J.; Yaver, D.; Hjort, C.M. Strategies and Challenges for the Development of Industrial Enzymes Using Fungal Cell Factories. In Grand Challenges in Biology and Biotechnology; Nevalainen, H., Ed.; Springer: Cham, Switzerland, 2020; pp. 179–210. ISBN 9783030295417. [Google Scholar]
- Biagini, R.; MacKenzie, B.; Sammons, D.; Smith, J.; Striley, C.; Robertson, S.; Snawder, J. Evaluation of the prevalence of anti-wheat-, anti—flour dust-, and anti-alpha amylase-specific IgE antibodies in blood donors. J. Allergy Clin. Immunol. 2003, 111, S95. [Google Scholar] [CrossRef]
- Green, B.J.; Beezhold, D.H. Industrial Fungal Enzymes: An Occupational Allergen Perspective. J. Allergy 2011, 2011, 682574. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, P.H.; Oxenbøll, K.M.; Wenzel, H. Cradle-to-Gate Environmental Assessment of Enzyme Products Produced Industrially in Denmark by Novozymes A/S. Int. J. Life Cycle Assess. 2007, 12, 432–438. [Google Scholar] [CrossRef]
- Sutay Kocabaş, D.; Grumet, R. Evolving regulatory policies regarding food enzymes produced by recombinant microorganisms. GM Crop. Food 2019, 10, 191–207. [Google Scholar] [CrossRef] [PubMed]
- Nikkilä, I.; Waldén, M.; Maina, N.H.; Tenkanen, M.; Mikkonen, K.S. Fungal Cell Biomass from Enzyme Industry as a Sustainable Source of Hydrocolloids. Front. Chem. Eng. 2020, 2, 1–12. [Google Scholar] [CrossRef]
- Owaid, M.N.; Abed, I.A.; Al-Saeedi, S.S.S. Applicable properties of the bio-fertilizer spent mushroom substrate in organic systems as a byproduct from the cultivation of Pleurotus spp. Inf. Process. Agric. 2017, 4, 78–82. [Google Scholar] [CrossRef]
- Ayele, A.; Haile, S.; Alemu, D.; Kamaraj, M. Comparative Utilization of Dead and Live Fungal Biomass for the Removal of Heavy Metal: A Concise Review. Sci. World J. 2021, 2021, 5588111. [Google Scholar] [CrossRef]
- Phan, C.-W.; Sabaratnam, V. Potential uses of spent mushroom substrate and its associated lignocellulosic enzymes. Appl. Microbiol. Biotechnol. 2012, 96, 863–873. [Google Scholar] [CrossRef]
- Rajavat, A.S.; Rai, S.; Pandiyan, K.; Kushwaha, P.; Choudhary, P.; Kumar, M.; Chakdar, H.; Singh, A.; Karthikeyan, N.; Bagul, S.Y.; et al. Sustainable use of the spent mushroom substrate ofPleurotus floridafor production of lignocellulolytic enzymes. J. Basic Microbiol. 2020, 60, 173–184. [Google Scholar] [CrossRef]
- Grujić, M.; Dojnov, B.; Potočnik, I.; Duduk, B.; Vujčić, Z. Spent mushroom compost as substrate for the production of industrially important hydrolytic enzymes by fungi Trichoderma spp. and Aspergillus niger in solid state fermentation. Int. Biodeterior. Biodegrad. 2015, 104, 290–298. [Google Scholar] [CrossRef]





| Application | Field | Fungal Name | Enzyme Name | Enzyme Use | Ref. |
|---|---|---|---|---|---|
| Industries | Food and beverage | Aspergillus oryzae, Aspergillus oryzae CCT 3940, and Fusariumculmorum ASP-87 | l-asparaginases | Reduce the acrylamide formation in potato chips or French fries, bakery products, and coffee by degradation of l-asparagine | [7,8,9] |
| Myceliophthora thermophilia | Laccases | Dough conditioner | [10] | ||
| Aspergillus niger DFR-5 | Xylanases | Improve yield and clarity of pineapple juice | [11] | ||
| Aspergillus niger | Pectinases | Improve the quantity of the extracted Orange juice | [12] | ||
| Talaromyces leycettanus | Pectinases | Efficiency in pectin degradation from grape juice | [13] | ||
| Pulp and paper | Pycnoporus cinnabarinus | Laccases | Improve the brightness and strength properties of the pulp | [14] | |
| Trametes villosa | Laccases | Internal sizing of paper by use of laccase and hydrophobic compounds | [15] | ||
| Trichoderma reesei QM9414 | Xylanases | Eco-friendly of biobleaching of Kraft pulp of sugarcane straw | [16] | ||
| Trichoderma viride VKF-3, Fusariumequiseti MF-3, and Aspergillus japonicus MF-1 | Cellulases, xylanases, laccases, and lipases | Treatment enhances the brightness, deinking and reduces the heavy metals in the newspaper pulp | [17] | ||
| Rhizopus oryzae MUCL 28168 and Fusarium solani | Tannases | Detoxification of coffee pulp by reduction of caffeine and tannins | [18] | ||
| Aspergillus niger, Phanerochaete chrysosporium, and Pycnoporus cinnabarinus | Feruloyl esterases, Mn2+-oxidizing peroxidases, and laccases | Decrease the final lignin content of flax pulp and improvement of pulp brightness | [19] | ||
| Textile | Aspergillus niger CKB and Trichoderma reesei ATCC 24449 | Cellulases | Textile waste hydrolysis for recovery of glucose and polyester | [20,21] | |
| Trichoderma longibrachiatum KT693225 | Xylanases | Desizing, bioscouring, and biofinishing of cellulosic fabrics (textile) without adding any additives | [22] | ||
| Aspergillus | Amylases | Desizing of cotton fibers by removal of starch from the surface of textile fibers | [23] | ||
| Candida orthopsilosis | Pectinases | Bioscouring of cotton fibers | [24] | ||
| Aspergillus niger and Penicillium | Glucose oxidases and catalases | Removal of hydrogen peroxide from cotton bioprocessing | [25] | ||
| Chaetomium globosum IMA1 | Lignin peroxidases laccases and manganese peroxidases | Decolorization of the industrial textile effluent | [26] | ||
| Environment | Biodegradation | Irpex lacteus and Pleurotusostreatus | Manganese-peroxidases and laccases | Biodegradation of chlorhexidine and octenidine as antimicrobial compounds used in oral careproducts | [27] |
| Marasmius sp. | Laccases | Degrade lignin by oxidizing the phenolic and non-phenolic compounds to produce dimers, oligomers, and polymers | [28] | ||
| Mucor circinelloides | Lipases, laccases, and peroxidases | Biodegradation of diesel oil hydrocarbons | [29] | ||
| Bioremediation | Penicillium sp. | Enzymatic reduction by the mer operon | The fungal enzyme could detoxify mercury (II) by extracellular sequestration via adsorption and precipitation | [30] | |
| Coriolopsis gallica | Laccases | Bioremediation of pollutants such as bisphenol, diclofenac, and 17-a-ethinylestradiol in real samples from the AQUIRIS wastewater | [31] | ||
| Aspergillus flavus FS4 and Aspergillus fumigates FS6 | Extracellular enzymes | Fungal consortium used for removal of chromium and cadmium | [32] | ||
| Pycnoporus sanguineus | Laccases | Fugal laccase was immobilized on calcium and copper alginate/chitosan beads and used for the removal of 17 a-ethinylestradiol | [33] | ||
| Thermomyces lanuginosus | Chitinases | Biocontrol agent against larvae of Eldana saccharina and fungi of Aspergillus sp., Mucor sp., and Fusarium verticillioides | [34] | ||
| Decolorization | Phanerochaete chrysosporium CDBB 686 | Ligninolytic enzymes | Decolorization of Congo red, Poly R-478, and Methyl green | [35] | |
| Geotrichum candidum | Peroxidases and laccases | Decolorization of methyl orange, Congo red, trypan blue, and Eriochrome black T | [36] | ||
| Coprinopsis cinerea | Laccases | High indigo dye decolorization | [37] | ||
| Trametes sp. SYBC-L4 | Laccases | Decolorization of Congo red, aniline blue, and indigo carmine | [38] | ||
| Phanerochaete chrysosporium | Manganese peroxidase | Decolorization of AO7 or CV pigment | [39] | ||
| Biomedical | Antimicrobial | 32 different isolated fungi identified by morphological characteristics and internal transcribed spacer sequence analysis | Amylases, proteases, pectinases, xylanases, cellulases, chitinases, and lipases | Antimicrobial activity against pathogenic organisms by agar diffusion assays | [40] |
| Aspergillus oryzae and Aspergillus flavipes | Proteases | Production of bioactive peptides from bovine and goat milk and the generated peptides tested against bacteria and fungi | [41] | ||
| Trichoderma harzianum | Chitinases | Degradation of chitosan to form chitosan-oligosaccharides and used as antimicrobial against pathogenic organisms | [42] | ||
| Anticancer | Trichoderma viride AUMC 13021 | Chitinases | Antitumor efficiency of chitinase against different types of cancer cell line | [43] | |
| Trichoderma harzianum | Chitinases | Chitosan-oligosaccharides used as anticancer compounds, which inhibited the growth of cervical cancer cells at concentration of 4 mg/mL and significantly reduced the survival rate of the cells | [42] | ||
| Aspergillus terreus | l-asparaginases | The synthesized zinc oxide conjugated l-asparaginase nanobiocomposite on MCF-7 cell line using MTT assay | [44] | ||
| Antioxidant | Aspergillus flavus | Catalases | Antioxidant system plays a crucial role in fungal development, aflatoxins biosynthesis, and virulence | [45] | |
| Pleurotus columbinus, P. foridanus, Aspergillus fumigatus, and Paecilomyces variotii | Peroxidases and catalases | Production of enzymatic antioxidant from peels of banana, pomegranate, and orange | [46] | ||
| Chytridiomycetes sp. | Ligninases | During biodegradation of lignin, the fungi synthesize bioactive compounds such as mycophenolic acid, dicerandrol C, phenyl acetates, anthrax quinones, benzo furans, and alkenyl phenols that have antioxidant activities | [47,48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Gendi, H.; Saleh, A.K.; Badierah, R.; Redwan, E.M.; El-Maradny, Y.A.; El-Fakharany, E.M. A Comprehensive Insight into Fungal Enzymes: Structure, Classification, and Their Role in Mankind’s Challenges. J. Fungi 2022, 8, 23. https://doi.org/10.3390/jof8010023
El-Gendi H, Saleh AK, Badierah R, Redwan EM, El-Maradny YA, El-Fakharany EM. A Comprehensive Insight into Fungal Enzymes: Structure, Classification, and Their Role in Mankind’s Challenges. Journal of Fungi. 2022; 8(1):23. https://doi.org/10.3390/jof8010023
Chicago/Turabian StyleEl-Gendi, Hamada, Ahmed K. Saleh, Raied Badierah, Elrashdy M. Redwan, Yousra A. El-Maradny, and Esmail M. El-Fakharany. 2022. "A Comprehensive Insight into Fungal Enzymes: Structure, Classification, and Their Role in Mankind’s Challenges" Journal of Fungi 8, no. 1: 23. https://doi.org/10.3390/jof8010023
APA StyleEl-Gendi, H., Saleh, A. K., Badierah, R., Redwan, E. M., El-Maradny, Y. A., & El-Fakharany, E. M. (2022). A Comprehensive Insight into Fungal Enzymes: Structure, Classification, and Their Role in Mankind’s Challenges. Journal of Fungi, 8(1), 23. https://doi.org/10.3390/jof8010023