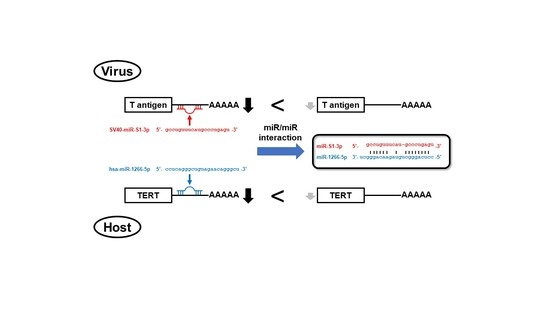
SV40 miR-S1 and Cellular miR-1266 Sequester Each Other from Their Targets, Enhancing Telomerase Activity and Viral Replication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Mammalian Expression Vectors
2.3. Retroviral Infection
2.4. Luciferase Reporter Assay
2.5. Pull-Down Analysis Using the Avidin–Biotin Binding System
2.6. Immunoprecipitation (IP) by Anti-Ago1, Ago2, Ago3, and Ago4 Antibodies
2.7. Quantitative Real-Time RT-PCR (RT-qPCR)
2.8. Semiquantitative RT-PCR
2.9. Immunoblotting
2.10. Telomeric Repeat Amplification Protocol (TRAP) Assay
2.11. Viral Infection Experiments
2.12. SV40 and SV40/TAD Preparation
2.13. Statistical Analysis
3. Results
3.1. MiR-S1-3p Directly Binds to Cellular miR-1266-5p
3.2. miR-S1-3p Prevents miR-1266-5p from Downregulating TERT
3.3. SV40 Infection Induced Mutual Sequestration between Viral miR-S1 and Cellular miR-1266 to Derepress Their Respective Targets, Telomerase and T Antigens
3.4. SV40 Infection-Induced miR-S1–miR-1266 Interplay Enhanced Telomerase Activity and Viral Replication
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, J.; DeCaprio, J.A.; Fluck, M.M.; Schaffhausen, B.S. Cellular transformation by simian virus 40 and murine polyoma virus T antigens. Semin. Cancer Biol. 2009, 19, 218–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, C.S.; Pipas, J.M. T antigens of simian virus 40: Molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 2002, 66, 179–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pipas, J.M. SV40: Cell transformation and tumorigenesis. Virology 2009, 384, 294–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.I.; Lickteig, R.L.; Estes, R.; Rundell, K.; Walter, G.; Munby, M.C. Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol. Cell Biol. 1991, 11, 1988–1995. [Google Scholar] [PubMed] [Green Version]
- Sontag, E.; Fedorov, S.; Kamibayashi, C.; Robbins, D.; Cobb, M.; Munby, M. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation. Cell 1993, 75, 887–897. [Google Scholar] [CrossRef]
- Chen, W.; Possemato, R.; Campbell, K.T.; Plattner, C.A.; Pallas, D.C.; Hahn, W.C. Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell 2004, 5, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Sontag, J.M.; Sontag, E. Regulation of cell adhesion by PP2A and SV40 small tumor antigen: An important link to cell transformation. Cell Mol. Life Sci. 2006, 63, 2979–2991. [Google Scholar] [CrossRef]
- Sullivan, C.S.; Grundhoff, A.T.; Tevethia, S.; Pipas, J.M.; Ganem, D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 2005, 435, 682–686. [Google Scholar] [CrossRef]
- Imperiale, M.J. Polyomavirus miRNAs: The beginning. Curr. Opin. Virol. 2014, 7, 29–32. [Google Scholar] [CrossRef] [Green Version]
- Tycowski, K.T.; Guo, Y.E.; Lee, N.; Moss, W.N.; Vallery, T.K.; Xie, M.; Steitz, J.A. Viral noncoding RNAs: More surprises. Genes Dev. 2015, 29, 567–584. [Google Scholar] [CrossRef] [Green Version]
- Gallo, A.; Bulati, M.; Miceli, V.; Amodia, N.; Conaldi, P.G. Non-coding RNAs: Strategy for viruses’ offensive. Non-Coding RNA 2020, 6, 38. [Google Scholar] [CrossRef]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [Green Version]
- Cazalla, D.; Yario, T.; Steitz, J.A. Down-regulation of a host microRNA by a herpesvirus saimiri noncoding RNA. Science 2010, 328, 1563–1566. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Shuai, M.; Xia, Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag. Res. 2019, 11, 8023–8031. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.S.; Su, J.L.; Cha, S.T.; Tarn, W.Y.; Wang, M.Y.; Hsu, H.C.; Lin, M.T.; Chu, C.Y.; Hua, K.T.; Chen, C.N.; et al. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J. Clin. Investig. 2011, 121, 3442–3455. [Google Scholar] [CrossRef] [Green Version]
- Tokorodani, M.; Ichikawa, H.; Yuasa, K.; Takahashi, T.; Hijikata, T. SV40 microRNA miR-S1-3p downregulates the expression of T antigens to control viral DNA replication, and TNFα and IL-17F expression. Biol. Pharm. Bull. 2020, 43, 1715–1728. [Google Scholar] [CrossRef]
- Chen, L.; Lu, M.H.; Zhang, D.; Hao, N.B.; Fan, Y.H.; Wu, Y.Y.; Wang, S.M.; Xie, R.; Fang, D.C.; Zhang, H.; et al. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014, 5, e1034. [Google Scholar] [CrossRef] [Green Version]
- Broekema, N.M.; Imperiale, M.J. miRNA regulation of BK polyomavirus replication during early infection. Proc. Natl. Acad. Sci. USA 2013, 110, 8200–8205. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.; Veldman, T.; Rundell, K.; Schlegel, R. Simian virus 40 small tumor antigen activates AKT and telomerase and induces anchorage-independent growth of human epithelial cells. J. Virol. 2002, 76, 10685–10691. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhao, L.L.; Funder, J.W.; Liu, J.P. Protein phosphatase 2A inhibits nuclear telomerase activity in human breast cancer cells. J. Biol. Chem. 1997, 272, 16729–16732. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, Y.; Cao, W.; Wang, C.; Sun, B.; Chen, J.; Li, S.; Chen, J.; Cui, M.; Zhang, B.; et al. miR-138-5p acts as a tumor suppressor by targeting hTERT in human colorectal cancer. Int. J. Exp. Pathol. 2017, 10, 11516–11525. [Google Scholar]
- Choy, E.Y.W.; Siu, K.L.; Kok, K.H.; Lung, R.W.M.; Tsang, C.M.; To, K.F.; Kwong, D.L.W.; Tsao, S.W.; Jin, D.Y. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 2008, 205, 2551–2560. [Google Scholar] [CrossRef] [Green Version]
- Gottwein, E.; Corcoran, D.L.; Mukherjee, N.; Skalsky, R.L.; Hafner, M.; Nusbaum, J.D.; Shamulailatpam, P.; Love, C.L.; Dave, S.S.; Tuschl, T.; et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 2011, 10, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Marquitz, A.R.; Mathur, A.; Shair, K.H.Y.; Raab-Traub, N. Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc. Natl. Acad. Sci. USA 2012, 109, 9593–9598. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Seo, D.; Kim, D.; Hong, Y.; Chang, H.; Baek, D.; Kim, V.N.; Lee, S.; Ahn, K. Temporal landscape of microRNA-mediated host-virus crosstalk during productive human cytomegalovirus infection. Cell Host Microbe 2015, 17, 838–851. [Google Scholar] [CrossRef] [Green Version]
- Qi, M.; Qi, Y.; Ma, Y.; He, R.; Ji, Y.; Sun, Z.; Ruan, Q. Over-expression of human cytomegalovirus miR-US25-2-3p downregulates eIF4A1 and inhibits HCMV replication. FEBS Lett. 2013, 587, 2266–2271. [Google Scholar] [CrossRef] [Green Version]
- Vojtechova, Z.; Tachezy, R. The role of miRNAs in virus-mediated oncogenesis. Int. J. Mol. Sci. 2018, 19, 1217. [Google Scholar] [CrossRef] [Green Version]
- Lo, A.K.F.; To, K.F.; Lo, K.W.; Lung, R.W.M.; Hui, J.W.Y.; Liao, G.; Hayward, S.D. Modulation of LMP1 protein exprtession by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. USA 2007, 104, 16164–16169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.T.; Zheng, H.B.; Zhang, D.Q.; Zhou, L.; Sun, H. MicroRNA-1266 suppresses papillary thyroid carcinoma cell metastasis and growth via targeting FGFR. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3430–3438. [Google Scholar]
- Su, Y.; Xie, R.; Xu, Q. Upregulation of miR-1266-5p serves as a prognostic biomarker of hepatocellular carcinoma and facilitates tumor cell proliferation, migration and invasion. Acta Biochim. Pol. 2021, 68, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Wang, X.; Li, J.; Wei, J.; Wang, Y.; Song, W.; Zhang, Z. MiR-1266 promotes cell proliferation, migration and invasion in cervical cancer by targeting DAB2IP. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3623–3630. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yu, H.; Yi, S.; Peng, X.; Su, P.; Xiao, Z.; Liu, R.; Tang, A.; Li, X.; Liu, F.; et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 2016, 7, 45370–45384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Xiong, Y.; Wu, Z.; He, Y.; Gao, X.; Zhou, Z.; Wang, T. MIR-138-5p inhibits the progression of prostate cancer by targeting FOXC1. Mol. Genet. Genom. Med. 2020, 8, e1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, W.; Li, R.; Liu, L.; Ni, X.; Shi, Y.; Xia, Y.; Wang, J.; Lu, F.; Xu, B. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote malignant melanoma progression by sponging miR-152-3p. Oncotarget 2017, 8, 85401–85414. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.; Wang, D.; Kong, Q.; Gao, W.; Sun, J. Function of miR-152 as a tumor suppressor in human breast cancer by targeting PIK3CA. Oncol. Res. 2017, 25, 1363–1371. [Google Scholar] [CrossRef]
- Ostadrahimi, S.; Valugerdi, M.A.; Hassan, M.; Haddad, G.; Fayaz, S.; Parvizhamidi, M.; Mahdian, R.; Esfahani, P.F. miR-1266-5p and miR-185-5p promote cell apoptosis in human prostate cancer cell lines. Asian Pac. J. Cancer Prev. 2018, 19, 2305–2311. [Google Scholar]
- Zhang, D.; Liu, X.; Zhang, Q.; Chen, X. miR-138-5p inhibits the malignant progression of prostate cancer by targeting FOXC1. Cancer Cell Int. 2020, 20, 297. [Google Scholar] [CrossRef]
- Zhang, W.; Liao, K.; Liu, D. MiR-138-5p inhibits the proliferation of gastric cancer cells by targeting DEK. Cancer Manag. Res. 2020, 12, 8137–8147. [Google Scholar] [CrossRef]
- Zheng, G.F.; Guo, N.H.; Zi, F.M.; Cheng, J. Long noncoding RNA H19 promotes tumorigenesis of multiple myeloma by activating BRD4 signaling by targeting microRNA 152-3p. Mol. Cell Biol. 2020, 40, e00382-19. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, J.; Zhang, Q.; Xu, H.; Liu, L. Long non-coding RNA HOXA11-AS modulates proliferation, apoptosis, metastasis and EMT in cutaneous melanoma cells partly via miR-152-3p/ITGA9 axis. Cancer Manag. Res. 2021, 13, 925–939. [Google Scholar] [CrossRef]
- Liu, X.; Li, L.; Bai, J.; Li, L.; Fan, J.; Fu, Z.; Liu, J. Long noncoding RNA plasmacytoma variant translocation 1 promotes progression of colorectal cancer by sponging microRNA-152-3p and regulating E2F3/MAPK8 signaling. Cancer Sci. 2022, 113, 109–119. [Google Scholar] [CrossRef]
- Lai, S.W.; Chen, M.Y.; Bamodu, O.A.; Hsieh, M.S.; Huang, T.Y.; Yeh, C.T.; Lee, W.H.; Cherng, Y.G. Exosomal lncRNA PVT1/VEGFA axis promotes colon cancer metastasis and stemness by downregulation of tumor suppressor miR-152-3p. Oxid. Med. Cell Longev. 2021, 2021, 9959807. [Google Scholar] [CrossRef]
- Barth, S.; Pfuhl, T.; Mamiani, A.; Ehses, C.; Roemer, K.; Kremmer, E.; Jäker, C.; Höck, J.; Meister, G.; Grässer, F.A. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF. Nucleic Acids Res. 2008, 36, 666–675. [Google Scholar] [CrossRef]
- Iizasa, H.; Wulff, B.E.; Alla, N.R.; Maragkakis, M.; Megraw, M.; Hatzigeorgiou, A.; Iwakiri, D.; Takada, K.; Wiedmer, A.; Showe, L.; et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and concequently affects viral latency. J. Biol. Chem. 2010, 285, 33358–33370. [Google Scholar] [CrossRef] [Green Version]
- Bellare, P.; Ganem, D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: An evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 2009, 6, 570–575. [Google Scholar] [CrossRef] [Green Version]
- Grey, F.; Meyers, H.; White, E.A.; Spector, D.H.; Nelson, J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007, 3, e163. [Google Scholar] [CrossRef]
- Murohy, E.; Vanicek, J.; Robins, H.; Shenk, T.; Levine, A. Suppression of immediate-early gene expression by herpesvirus-coded microRNAs: Implications for latency. Proc. Natl. Acad. Sci. USA 2008, 105, 5453–5458. [Google Scholar] [CrossRef] [Green Version]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.J.; Choi, H.; Kim, H.; Lee, S.K. MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF. J. Virol. 2014, 88, 9027–9037. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.C.; Li, Z.; Chu, C.Y.; Feng, J.; Feng, J.; Sun, R.; Rana, T.M. MicroRNAs encoded by Kaposi’s sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. 2010, 11, 784–790. [Google Scholar] [CrossRef] [Green Version]
- Brown, J. Herpes simplex virus latency: The DNA repair-centered pathway. Adv. Virol. 2017, 2017, 7028194. [Google Scholar] [CrossRef]
- Kenney, S.; Mertz, J.E. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin. Cancer Biol. 2014, 26, 60–68. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, T.; Ichikawa, H.; Okayama, Y.; Seki, M.; Hijikata, T. SV40 miR-S1 and Cellular miR-1266 Sequester Each Other from Their Targets, Enhancing Telomerase Activity and Viral Replication. Non-Coding RNA 2022, 8, 57. https://doi.org/10.3390/ncrna8040057
Takahashi T, Ichikawa H, Okayama Y, Seki M, Hijikata T. SV40 miR-S1 and Cellular miR-1266 Sequester Each Other from Their Targets, Enhancing Telomerase Activity and Viral Replication. Non-Coding RNA. 2022; 8(4):57. https://doi.org/10.3390/ncrna8040057
Chicago/Turabian StyleTakahashi, Tetsuyuki, Hirona Ichikawa, Yukiko Okayama, Manami Seki, and Takao Hijikata. 2022. "SV40 miR-S1 and Cellular miR-1266 Sequester Each Other from Their Targets, Enhancing Telomerase Activity and Viral Replication" Non-Coding RNA 8, no. 4: 57. https://doi.org/10.3390/ncrna8040057
APA StyleTakahashi, T., Ichikawa, H., Okayama, Y., Seki, M., & Hijikata, T. (2022). SV40 miR-S1 and Cellular miR-1266 Sequester Each Other from Their Targets, Enhancing Telomerase Activity and Viral Replication. Non-Coding RNA, 8(4), 57. https://doi.org/10.3390/ncrna8040057