Transcription Factors Controlling Primary and Secondary Metabolism in Filamentous Fungi: The β-Lactam Paradigm
Abstract
:1. Introduction
2. Zinc-Binding Transcription Factors
2.1. Class I Zinc Binding Transcription Factors (C2H2)
2.1.1. Class I: CreA (Cre1)
2.1.2. Class I: PacC
2.1.3. Class I: MTFA (Master Transcription Factor A)
2.2. Class II Zinc Binding Transcription Factors (C4)
Class II: AreA (Nre)
2.3. Class III Zinc Binding Transcription Factors (Zn2C6)
Specificity and Binding of the Binuclear Zinc Finger Transcription Factors
3. Winged Helix Regulators
3.1. RFX-Related Transcription Regulators
3.2. Forkhead-Type Regulators
4. Global Regulators of Secondary Metabolism
4.1. The LaeA Transcription Factor and Heterochromatin Reorganization
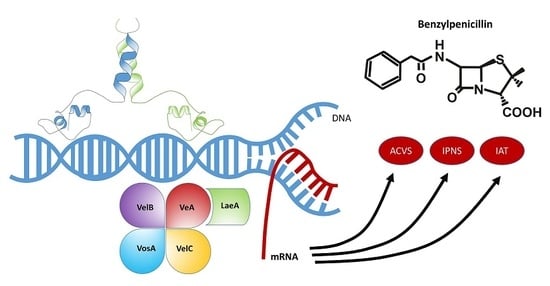
4.2. The Velvet Complex
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnston, S.R.; Boddy, L.; Weightman, A.J. Bacteria in decomposing wood and their interactions with wood-decay fungi. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [Green Version]
- Janusz, G.; Pawlik, A.; Sulej, J.; Swiderska-Burek, U.; Jarosz-Wilkolazka, A.; Paszczynski, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Liu, N.; Li, C.; Wang, X.; Xu, X.; Chen, W.; Xing, G.; Zheng, W. The early response during the interaction of fungal phytopathogen and host plant. Open Biol. 2017, 7, 170057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickie, I.A.; Bufford, J.L.; Cobb, R.C.; Desprez-Loustau, M.L.; Grelet, G.; Hulme, P.E.; Klironomos, J.; Makiola, A.; Nuñez, M.A.; Pringle, A.; et al. The emerging science of linked plant-fungal invasions. New Phytol. 2017, 215, 1314–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karathia, H.; Vilaprinyo, E.; Sorribas, A.; Alves, R. Saccharomyces cerevisiae as a model organism: A comparative study. PLoS ONE 2011, 6, e16015. [Google Scholar] [CrossRef] [PubMed]
- Adrio, J.L.; Demain, A.L. Fungal Biotechnology. Int. Microbiol. 2003, 6, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Valuable Secondary Metabolites from fungi. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Martín, J.F., García-Estrada, C., Zeilinger, S., Eds.; Springer Verlag: New York, NY, USA, 2014; Volume 1, pp. 1–15. ISBN 978-1-4939-1190-5. [Google Scholar]
- Zeillinger, S.; García-Estrada, C.; Martin, J.F. Fungal Secondary metabolites in the OMICS Era. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Zeilinger, S., Martín, J.F., García-Estrada, C., Eds.; Springer Verlag: New York, NY, USA, 2015; Volume 2, pp. 1–12. ISBN 978-1-4939-5516-9. [Google Scholar]
- Van den Berg, M.A.; Albang, R.; Albermann, K.; Badger, J.H.; Daran, J.M.; Driessen, A.J.; Garcia-Estrada, C.; Fedorova, N.D.; Harris, D.M.; Heijne, W.H.; et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 2008, 26, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H.; Keller, N. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 2005, 43, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Laich, F.; Fierro, F.; Martín, J.F. Production of penicillin by fungi growing on food products: Identification of a complete penicillin gene cluster in Penicillium griseofulvum and a truncated cluster in Penicillium verrucosum. Appl. Environ. Microbiol. 2002, 68, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Ullán, R.V.; García-Estrada, C. Regulation and compartmentalization of β-lactam biosynthesis. Microb. Biotechnol. 2010, 3, 285–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seetharam, A.; Stuart, G.W. A study on the distribution of 37 well conserved families of C2H2 zinc finger genes in eukaryotes. BMC Genom. 2013, 14, 420. [Google Scholar] [CrossRef] [PubMed]
- Malgieri, G.; Palmieri, M.; Russo, L.; Fattorusso, R.; Pedone, P.V.; Isernia, C. The prokaryotic zinc-finger: Structure, function and comparison with the eukaryotic counterpart. FEBS J. 2015, 282, 4480–4496. [Google Scholar] [CrossRef] [PubMed]
- MacPherson, S.; Larochelle, M.; Turcotte, B. A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006, 70, 583–604. [Google Scholar] [CrossRef] [PubMed]
- Siggers, T.; Reddy, J.; Barron, B.; Bulyk, M.L. Diversification of transcription factor paralogs via noncanonical modularity in C2H2 zinc finger DNA binding. Mol. Cell 2014, 55, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Jekosch, K.; Kück, U. Glucose dependent transcriptional expression of the cre1 gene in Acremonium chrysogenum strains showing different levels of cephalosporin C production. Curr. Genet. 2000, 37, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Jekosch, K.; Kück, U. Loss of glucose repression in an Acremonium chrysogenum beta-lactam producer strain and its restoration by multiple copies of the cre1 gene. Appl. Microbiol. Biotechnol. 2000, 54, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Espeso, E.A.; Peñalva, M.A. Three binding sites for the Aspergillus nidulans PacC zinc-finger transcription factor are necessary and sufficient for regulation by ambient pH of the isopenicillin N synthase gene promoter. J. Biol. Chem. 1996, 271, 28825–28830. [Google Scholar] [CrossRef] [PubMed]
- Park, D.S.; Yu, Y.M.; Kim, Y.J.; Maeng, P.J. Negative regulation of the vacuole-mediated resistance to K(+) stress by a novel C2H2 zinc finger transcription factor encoded by aslA in Aspergillus nidulans. J. Microbiol. 2015, 53, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Shantappa, S.; Dhingra, S.; Hernández-Ortiz, P.; Espeso, E.A.; Calvo, A.M. Role of the zinc finger transcription factor SltA in morphogenesis and sterigmatocystin biosynthesis in the fungus Aspergillus nidulans. PLoS ONE 2013, 8, e68492. [Google Scholar] [CrossRef] [PubMed]
- Kwon, N.J.; Garzia, A.; Espeso, E.A.; Ugalde, U.; Yu, J.H. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 2010, 77, 1203–1219. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Chae, K.S.; Han, K.H.; Han, D.M. The nsdC gene encoding a putative C2H2-type transcription factor is a key activator of sexual development in Aspergillus nidulans. Genetics 2009, 182, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, V.; Dhingra, S.; Kincaid, A.; Shantappa, S.; Feng, X.; Calvo, A.M. The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS ONE 2013, 8, e74122. [Google Scholar] [CrossRef] [PubMed]
- Adams, T.H.; Wieser, J.K.; Yu, J.H. Asexual Sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 1998, 62, 35–54. [Google Scholar] [PubMed]
- Dowzer, C.E.; Kelly, J.M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 1991, 11, 5701–5709. [Google Scholar] [CrossRef] [PubMed]
- Espeso, E.A.; Peñalva, M.A. Carbon catabolite repression can account for the temporal pattern of expression of a penicillin biosynthetic gene in Aspergillus nidulans. Mol. Microbiol. 1992, 6, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Ren, Y.; Zhou, X.; Cai, M.; Zhang, Y. Transcription factor Agseb1 affects development, osmotic stress response, and secondary metabolism in marine-derived Aspergillus glaucus. J. Basic Microbiol. 2017, 57, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.; Dalmais, B.; Morgant, G.; Viaud, M. Screening of a Botrytis cinerea one-hybrid library reveals a Cys2His2 transcription factor involved in the regulation of secondary metabolism gene clusters. Fungal Genet. Biol. 2013, 52, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.K.; Barad, S.; Luria, N.; Kumar, D.; Espeso, E.A.; Prusky, D.B. Cation-stress-responsive transcription factors SltA and CrzA regulate morphogenetic processes and pathogenicity of Colletotrichum gloeosporioides. PLoS ONE 2016, 11, e0168561. [Google Scholar] [CrossRef] [PubMed]
- Malapi-Wight, M.; Kim, J.E.; Shim, W.B. The N-terminus region of the putative C2H2 transcription factor Ada1 harbors a species-specific activation motif that regulates asexual reproduction in Fusarium verticillioides. Fungal Genet. Biol. 2014, 62, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Malapi-Wight, M.; Smith, J.; Campbell, J.; Bluhm, B.H.; Shim, W.B. Sda1, a Cys2-His2 zinc finger transcription factor, is involved in polyol metabolism and fumonisin B1 production in Fusarium verticillioides. PLoS ONE 2013, 8, e67656. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Que, Y.; Xu, L.; Deng, S.; Peng, Y.; Talbot, N.J.; Wang, Z. ZNF1 encodes a putative C2H2 Zinc-finger protein essential for appressorium differentiation by the rice blast fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 2016, 29, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Matheis, S.; Yemelin, A.; Scheps, D.; Andresen, K.; Jacob, S.; Thines, E.; Foster, A.J. Functions of the Magnaporthe oryzae Flb3p and Flb4p transcription factors in the regulation of conidiation. Microbiol. Res. 2017, 196, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Han, X.; Liu, Z.; He, C. The function and properties of the transcriptional regulator COS1 in Magnaporthe oryzae. Fungal Biol. 2013, 117, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Nehlin, J.O.; Ronne, H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms’ tumour finger proteins. EMBO J. 1990, 15, 2891–2898. [Google Scholar]
- Cetz-Chel, J.E.; Balcázar-López, E.; Esquivel-Naranjo, E.U.; Herrera-Estrella, A. The Trichoderma atroviride putative transcription factor Blu7 controls light responsiveness and tolerance. BMC Genom. 2016, 17, 327. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Wang, Y.; Deng, C.; Hu, R.; Tian, C. Phylogenic analysis revealed an expanded C2H2-homeobox subfamily and expression profiles of C2H2 zinc finger gene family in Verticillium dahliae. Gene 2015, 562, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Pelkmans, J.F.; Vos, A.M.; Scholtmeijer, K.; Hendrix, E.; Baars, J.J.; Gehrmann, T.; Reinders, M.J.; Lugones, L.G.; Wösten, H.A. The transcriptional regulator c2h2 accelerates mushroom formation in Agaricus bisporus. Appl. Microbiol. Biotechnol. 2016, 100, 7151–7159. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Wright, L.C.; Tscharke, R.L.; Sorrell, T.C.; Wilson, C.F.; Kwon-Chung, K.J. Regulatory roles for the homeodomain and C2H2 zinc finger regions of Cryptococcus neoformans Ste12alphap. Mol. Microbiol. 2004, 53, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.L.; Zhang, G.; Ren, A.; Dang, Z.H.; Shi, L.; Jiang, A.L.; Zhao, M.W. The pH-responsive transcription factor PacC regulates mycelial growth, fruiting body development, and ganoderic acid biosynthesis in Ganoderma lucidum. Mycologia 2016, 108, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Ohm, R.A.; de Jong, J.F.; de Bekker, C.; Wösten, H.A.B.; Lugones, L.G. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol. Microbiol. 2011, 15, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Kief, J.; Auffarth, K.; Farfsing, J.W.; Mahlert, M.; Nieto, F.; Basse, C.W. The Ustilago maydis Cys2His2-type zinc finger transcription factor Mzr1 regulates fungal gene expression during the biotrophic growth stage. Mol. Microbiol. 2008, 68, 1450–1470. [Google Scholar] [CrossRef] [PubMed]
- Teichmann, B.; Liu, L.; Schink, K.O.; Bölker, M. Activation of the ustilagic acid biosynthesis gene cluster in Ustilago maydis by the C2H2 zinc finger transcription factor Rua1. Appl. Environ. Microbiol. 2010, 76, 2633–2640. [Google Scholar] [CrossRef] [PubMed]
- Kulmburg, P.; Judewicz, N.; Mathieu, N.; Lenouvel, F.; Sequeval, D.; Felenbok, B. Specific binding sites for the activator protein ALcR in the alcA promoter of the ethanol regulon in Aspergillus nidulans. J. Biol. Chem. 1992, 267, 21146–21153. [Google Scholar] [PubMed]
- Nikolaev, I.; Lenouvel, F.; Felenbok, B. Unique DNA binding specificity of the binuclear zinc AlcR activator of the ethanol utilization pathway in Aspergillus nidulans. J. Biol. Chem. 1999, 274, 9795–9802. [Google Scholar] [CrossRef] [PubMed]
- Empel, J.; Sitkiewicz, I.; Andrukiewicz, A.; Lasocki, K.; Borsuk, P.; Weglenski, P. arcC, the regulatory gene for the arginine catabolic pathway in Aspergillus nidulans. Mol. Gen. Genom. 2001, 266, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Burger, G.; Strauss, J.; Scazzocchio, C.; Lang, B.F. nirA, the pathway-specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative Gal4-type zinc finger protein and contains four introns in highly conserved regions. Mol. Cell. Biol. 1991, 11, 5746–5755. [Google Scholar] [CrossRef] [PubMed]
- Punt, P.J.; Strauss, J.; Smit, R.; Kinghorn, J.R.; van den Hondel, C.A.; Scazzocchio, C. The intergenic region between the divergently transcribed niaA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol. Cell. Biol. 1995, 15, 5688–5699. [Google Scholar] [CrossRef] [PubMed]
- Cazell, B.; Pokorska, A.; Hull, E.; Green, P.M.; Stanway, G.; Scazzocchio, C. Sequence, exon-intron organization transcription and mutational analysis of prnA, the gene encoding the transcriptional activator of the prn gene cluster in Aspergillus nidulans. Mol. Microbiol. 1998, 28, 355–370. [Google Scholar] [CrossRef]
- Gómez, D.; Cubero, B.; Cecchetto, G.; Scazzocchio, C. PrnA, a Zn(2)Cys(6) activator with a unique DNA recognition mode, requires inducer for in vivo binding. Mol. Microbiol. 2002, 44, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Levesley, I.; Newton, G.H.; Lamb, H.K.; Vanschothorst, E.; Dalgleish, R.W.M.; Samson, A.C.R.; Roberts, C.F.; Hawkins, A.R. Domain-structure and function within the QUTA protein of Aspergillus nidulans-implications for the control of transcription. Microbiology 1996, 142, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Lamb, H.K.; Roberts, C.F.; Hawkins, A.R. A second gene (qutH) within the Aspergillus nidulans quinic acid utilization gene cluster encodes a protein with a putative zinc-cluster motif. Gene 1992, 112, 219–224. [Google Scholar] [CrossRef]
- Davis, M.A.; Small, A.J.; Kourambas, S.; Hynes, M.J. The tamA gene of Aspergillus nidulans contains a putative zinc cluster motif which is not required for gene function. J. Bacteriol. 1996, 178, 3406–3409. [Google Scholar] [CrossRef] [PubMed]
- Ceccetto, G.; Amillis, S.; Diallinas, G.; Scazzochio, C.; Drevet, C. The AzgA purine transporter of Aspergillus nidulans. Characterization of a protein belonging to a new phylogenetic cluster. J. Biol. Chem. 2004, 279, 3132–3141. [Google Scholar] [CrossRef] [PubMed]
- Suárez, T.; Queiroz, M.V.; Oestreicher, N.; Scazzocchio, C. The sequence and binding specificity of UaY, the specific regulator of the purine utilization pathway in Aspergillus nidulans suggest an evolutionary relationship with the PPR1 of Saccharomyces cerevisiae. EMBO J. 1995, 14, 1453–1467. [Google Scholar] [PubMed]
- Gomi, K.; Akeno, T.; Minetoki, T.; Ozeki, K.; Kumagai, C.; Okazaki, N.; Iimura, Y. Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2000, 64, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Tani, S.; Katsuyama, Y.; Hayashi, T.; Suzuki, H.; Kato, M.; Gomi, K.; Kobayashi, T.; Tsukagoshi, N. Characterization of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr. Genet. 2001, 39, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.B.; Nurphy, R.L.; Martin, H.M.; Sharps, J.A.; Davis, M.A.; Katz, M.E.; Hynes, M.J. The acetate regulatory gene FacB of Aspergillus nidulans encodes a Zn(II)2Cys6 transcriptional activator. Mol. Gen. Genet. 1997, 254, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.B.; Andrianopoulos, A.; Davis, M.A.; Hynes, M.J. FacB, the Aspergillus nidulans activator of acetate utilization genes, binds dissimilar DNA sequences. EMBO J. 1998, 17, 2042–2054. [Google Scholar] [CrossRef] [PubMed]
- Hasper, A.A.; Visser, J.; Graaff, L.H. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates D-xylose reductase gene expression. Mol. Microbiol. 2000, 36, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, D.M.; Howlett, B.J. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol. Lett. 2005, 248, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Kihara, J.; Moriwaki, A.; Tanaka, N.; Tanaka, C.; Ueno, M.; Arase, S. Characterization of the BMR1 gene encoding a transcription factor for melanin biosynthesis genes in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 2008, 281, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Hao, T.; Wang, Y.; Pan, Y.; Ren, F.; Liu, X.; Che, Y.; Liu, G. VerZ, a Zn(II)2Cys6 DNA-binding protein, regulates the biosynthesis of verticillin in Clonostachys rogersoniana. Microbiology 2017, 163, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, M.; Perfect, S.; Pellier, A.L.; Bailey, J.A.; Langin, I. A Gal4-like protein is involved in the switch between biotrophic and necrotrophic phases of the infection process of Colletotrichum lindemuthianum on common vean. Plant Cell. 2000, 12, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, G.; Kenmochi, Y.; Takano, J.; Sweigard, J.; Farrall, L.; Furusawa, I.; Horino, O.; Kubo, Y. Novel fungal transcriptional activator, Cmr1p of Colletotrichum lagenarium and Pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmental manner. Mol. Microbiol. 2000, 38, 940–954. [Google Scholar] [PubMed]
- Chung, K.R.; Daub, M.E.; Kuchler, K.; Schuller, C. The CRG1 gene required for resistance to the singlet oxygen-generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem. Biophys. Res. Commun. 2003, 302, 302–310. [Google Scholar] [CrossRef]
- Flaherty, J.E.; Woloshuk, C.P. Regulation of the fumonisin biosynthesis in Fusarium verticilloides by a zinc binuclear cluster-type gene, ZFR1. Appl. Environ. Microbiol. 2004, 70, 2653–2659. [Google Scholar] [CrossRef] [PubMed]
- Imazaki, I.; Kurahashi, M.; Iida, Y.; Tsuge, T. Fow2, a Zn(II)2Cys6-type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum. Mol. Microbiol. 2007, 63, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.; Son, H.; Choi, G.J.; Lee, C.; Kim, J.C.; Kim, H.; Lee, Y.W. Transcription factor ART1 mediates starch hydrolysis and mycotoxin production in Fusarium graminearum and F. verticillioides. Mol. Plant Pathol. 2016, 17, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Waalwijk, C.; de Wit, P.J.; van der Lee, T.; Tang, D. EBR1, a novel Zn(2)Cys(6) transcription factor, affects virulence and apical dominance of the hyphal tip in Fusarium graminearum. Mol. Plant. Microbe Interact. 2011, 24, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.M.; Gardiner, D.M.; Keller, N.P.; Howlett, B.J. A Zn(II)2Cys6 DNA binding protein regulates the sirodesmin PL biosynthetic gene cluster in Leptosphaeria maculans. Fungal Genet. Biol. 2008, 45, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Li, D.X.; Kolattukudy, P.E. Cloning of cutinase transcription factor 1, a transactivating protein containing Cys(6)Zn(2) binuclear cluster DNA_binding motif. J. Biol. Chem. 1997, 272, 12462–12467. [Google Scholar] [CrossRef] [PubMed]
- Bailey, L.A.; Ebbole, D.J. The Fluffy gene of Neurospora crassa encodes a Gal4p-type C6 zinc cluster protein required for conidial development. Genetics 1998, 148, 1813–1820. [Google Scholar] [PubMed]
- Feng, B.; Marzluf, G.A. The regulatory protein NIT4 that mediates nitrate induction in Neurospora crassa contains a complex tripartite activation domain with a novel leucine-rich, acidic motif. Curr. Genet. 1996, 29, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Ono, C.; Hosobuchi, M.; Yoshikawa, H. Functional analysis of mlcR, a regulatory gene for ML-236B (compactin) biosynthesis in Penicillium citrinum. Mol. Genet. Genom. 2002, 268, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Gil-Duran, C.; Rojas-Aedo, J.F.; Medina, E.; Vaca, I.; García-Rico, R.O.; Villagrán, S.; Levicán, G.; Chávez, R. The pcz1 gene, which encodes a Zn(II)2Cys6 protein, is involved in the control of growth, conidiation, and conicidial germination in the filamentous fungus Penicillium roqueforti. PLoS ONE 2015, 10, e0120740. [Google Scholar] [CrossRef] [PubMed]
- Rybak, K.; See, P.T.; Phan, H.T.; Syme, R.A.; Moffat, C.S.; Oliver, R.P.; Tan, K.C. A functionally conserved Zn(2)Cys(6) binuclear cluster transcription factor class regulates necrotrophic effector gene expression and host-specific virulence of two major Pleosporales fungal pathogens of wheat. Mol. Plant. Pathol. 2017, 18, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Messenguy, F.; Dubois, E.; Descamps, F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur. J. Biochem. 1986, 15, 77–81. [Google Scholar] [CrossRef]
- Canovas, D.; Andrianopoulus, A. Developmental regulation of the glyoxalate cycle in the human pathogen Penicillium marneffei. Mol. Microbiol. 2006, 62, 1725–1738. [Google Scholar] [CrossRef] [PubMed]
- Aro, N.; Saloheimo, A.; Ilmen, M.; Pentilla, M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes in Trichoderma reesei. J. Biol. Chem. 2001, 276, 24309–24314. [Google Scholar] [CrossRef] [PubMed]
- Masloff, S.; Poggeler, S.; Kuck, U. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 1999, 152, 191–199. [Google Scholar] [PubMed]
- Ohm, R.A.; de Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; de Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E.; et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 15, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Horst, R.J.; Zeh, C.; Saur, A.; Sonnewald, S.; Sonnewald, U.; Voll, L.M. The Ustilago maydis Nit2 homolog regulates nitrogen utilization and is required for efficient induction of filamentous growth. Eukaryot Cell. 2012, 11, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Fedotova, A.A.; Bonchuk, A.N.; Mogila, V.A.; Georgiev, P.G. C2H2 Zinc finger proteins: The largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae 2017, 9, 47–58. [Google Scholar] [PubMed]
- Esperón, P.; Scazzocchio, C.; Paulino, M. In vitro and in silico analysis of the Aspergillus nidulans DNA-CreA repressor interactions. J. Biomol. Struct. Dyn. 2014, 32, 2033–2041. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, G.J.; Visser, J. Carbon repression in Aspergilli. FEMS Microbiol. Lett. 1997, 151, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Ronne, H. Glucose repression in fungi. Trends Genet. 1995, 11, 12–17. [Google Scholar] [CrossRef]
- Adnan, M.; Zheng, W.; Islam, W.; Arif, M.; Abubakar, Y.S.; Wang, Z.; Lu, G. Carbon catabolite repression in filamentous fungi. Int. J. Mol. Sci. 2017, 19, 48. [Google Scholar] [CrossRef] [PubMed]
- Cubero, B.; Scazzocchio, C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994, 13, 407–415. [Google Scholar] [PubMed]
- Cubero, B.; Gómez, D.; Scazzocchio, C. Metabolite repression and inducer exclusion in the proline utilization gene cluster of Aspergillus nidulans. J. Bacteriol. 2000, 182, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Kulmburg, P.; Mathieu, M.; Dowzer, C.; Kelly, J.; Felenbok, B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol. Microbiol. 1993, 7, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Revilla, G.; López-Nieto, M.J.; Luengo, J.M.; Martín, J.F. Carbon catabolite repression of penicillin biosynthesis by Penicillium chrysogenum. J. Antibiot. Tokyo 1984, 37, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A. Molecular Regulation of Beta-lactam Biosynthesis in Filamentous Fungi. Microbiol. Mol. Biol. Rev. 1998, 62, 547–585. [Google Scholar] [PubMed]
- Gutiérrez, S.; Fierro, F.; Casqueiro, J.; Martín, J.F. Gene organization and plasticity of the beta-lactam genes in different filamentous fungi. Antonie van Leeuwenhoek 1999, 75, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Casqueiro, J.; Kosalková, K.; Marcos, A.T.; Gutiérrez, S. Penicillin and cephalosporin biosynthesis: Mechanism of carbon catabolite regulation of penicillin production. Antonie van Leeuwenhoek 1999, 75, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Cepeda-García, C.; Domínguez-Santos, R.; García-Rico, R.O.; García-Estrada, C.; Cajiao, A.; Fierro, F.; Martín, J.F. Direct involvement of the CreA transcription factor in penicillin biosynthesis and expression of the pcbAB gene in Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2014, 98, 7113–7124. [Google Scholar] [CrossRef] [PubMed]
- Behmer, C.J.; Demain, A.L. Further studies on carbon catabolite regulation of β-lactam antibiotic synthesis in Cephalosporium acremonium. Curr. Microbiol. 1983, 8, 107–114. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Browne, P.; Turner, G. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J. Bacteriol. 1992, 174, 3789–3799. [Google Scholar] [CrossRef] [PubMed]
- Revilla, G.; Ramos, F.R.; López-Nieto, M.J.; Alvarez, E.; Martín, J.F. Glucose represses formation of delta-(l-alpha-aminoadipyl)-l-cysteinyl-d-valine and isopenicillin N synthase but not penicillin acyltransferase in Penicillium chrysogenum. J. Bacteriol. 1986, 168, 947–952. [Google Scholar] [CrossRef] [PubMed]
- Lockington, R.A.; Kelly, J.M. Carbon catabolite repression in Aspergillus nidulans involves deubiquitination. Mol. Microbiol. 2001, 40, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.B.; Lockington, R.A.; Kelly, J.M. The Aspergillus nidulans creC gene involved in carbon catabolite repression encodes a WD40 repeat protein. Mol. Gen. Genet. 2000, 263, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wolfe, S.; Demain, A.L. Carbon source regulation of the ACV synthetase in Cephalosporium acremonium C-10. Curr. Microbiol. 1989, 18, 361–367. [Google Scholar] [CrossRef]
- Tilburn, J.; Sarkar, S.; Widdick, D.A.; Espeso, E.A.; Orejas, M.; Mungroo, J.; Peñalva, M.A.; Arst, H.N., Jr. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995, 14, 779–790. [Google Scholar] [PubMed]
- Espeso, E.A.; Tilburn, J.; Sánchez-Pulido, L.; Brown, C.V.; Valencia, A.; Arst, H.N., Jr.; Peñalva, M.A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J. Mol. Biol. 1997, 274, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Espeso, E.A.; Roncal, T.; Díez, E.; Rainbow, L.; Bignell, E.; Alvaro, J.; Suárez, T.; Denison, S.H.; Tilburn, J.; Arst, H.N., Jr.; et al. On how a transcription factor can avoid its proteolytic activation in the absence of signal transduction. EMBO J. 2000, 19, 719–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peñalva, M.A.; Lucena-Agell, D.; Arst, H.N., Jr. Liaison alcaline: Pals entice non-endosomal ESCRTs to the plasma membrane for pH signaling. Curr. Opin. Microbiol. 2014, 22, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Bussink, H.J.; Bignell, E.M.; Múnera-Huertas, T.; Lucena-Agell, D.; Scazzocchio, C.; Espeso, E.A.; Bertuzzi, M.; Rudnicka, J.; Negrete-Urtasun, S.; Peñas-Parilla, M.M.; et al. Refining the pH response in Aspergillus nidulans: A modulatory triad involving PacX, a novel Zinc binuclear cluster protein. Mol. Microbiol. 2015, 98, 1051–1072. [Google Scholar] [CrossRef] [PubMed]
- Barwell, K.J.; Boysen, J.H.; Xu, W.; Mitchell, A.P. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 2005, 4, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Rothfels, K.; Tanny, J.C.; Molnar, E.; Friesen, H.; Commisso, C.; Segall, J. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 2005, 25, 6772–6788. [Google Scholar] [CrossRef] [PubMed]
- Calcagno-Pizarelli, A.M.; Negrete-Urtasun, S.; Denison, S.H.; Rudnicka, J.D.; Bussink, H.J.; Múnera-Huertas, T.; Stanton, L.; Hervás-Aguilar, A.; Espeso, E.A.; Tilburn, J.; et al. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot. Cell 2007, 6, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Herranz, S.; Rodríguez, J.M.; Bussink, H.J.; Sánchez-Ferrero, J.C.; Arst, H.N., Jr.; Peñalva, M.A.; Vincent, O. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. USA 2005, 102, 12141–12146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hervás-Aguilar, A.; Galindo, A.; Peñalva, M.A. Receptor-independent ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J. Biol. Chem. 2010, 285, 18095–18102. [Google Scholar] [CrossRef] [PubMed]
- Selvig, K.; Alspaugh, J.A. pH response pathways in fungi: Adapting to host-derived and environmental signals. Mycobiology 2011, 39, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Vincent, O.; Rainbow, L.; Tilburn, J.; Arst, H.N., Jr.; Peñalva, M.A. YPXL/I is a protein interaction motif recognized by aspergillus PalA and its human homologue, AIP1/Alix. Mol. Cell. Biol. 2003, 23, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Díez, E.; Alvaro, J.; Espeso, E.A.; Rainbow, L.; Suárez, T.; Tilburn, J.; Arst, H.N., Jr.; Peñalva, M.A. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002, 21, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Galán, O.; Galindo, A.; Hervás-Aguilar, A.; Arst, H.N., Jr.; Peñalva, M.A. Physiological involvement in pH signaling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J. Biol. Chem. 2009, 284, 4404–4412. [Google Scholar] [CrossRef] [PubMed]
- Lucena-Agell, D.; Galindo, A.; Arst, H.N., Jr.; Peñalva, M.A. Aspergillus nidulans Ambient pH signaling does not require endocytosis. Eukaryot. Cell 2015, 14, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Hervás-Aguilar, A.; Rodríguez, J.M.; Tilburn, J.; Arst, H.N., Jr.; Peñalva, M.A. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J. Biol. Chem. 2007, 282, 34735–34747. [Google Scholar] [CrossRef] [PubMed]
- Suárez, T.; Peñalva, M.A. Characterization of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol. Microbiol. 1996, 20, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, E.K.; Kempken, R.; Kück, U. Functional analysis of promoter sequences of cephalosporin C biosynthesis genes from Acremonium chrysogenum: Specific DNA-protein interactions and characterization of the transcription factor PACC. Mol. Genet. Genom. 2001, 265, 508–518. [Google Scholar]
- Espeso, E.A.; Tilburn, J.; Arst, H.N., Jr.; Peñalva, M.A. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 1993, 12, 3947–3956. [Google Scholar] [PubMed]
- Yager, L.N. Early developmental events during asexual and sexual sporulation in Aspergillus nidulans. Biotechnology 1992, 23, 19–41. [Google Scholar] [PubMed]
- Urnov, F.D. A feel for the template: Zinc finger protein transcription factors and chromatin. Biochem. Cell. Biol. 2002, 80, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Teakle, G.R.; Gilmartin, P.M. Two forms of type IV zinc-finger motif and their kingdom-specific distribution between the flora, fauna and fungi. Trends Biochem. Sci. 1998, 23, 100–102. [Google Scholar] [CrossRef]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insight into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Marzluf, G.A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 1997, 61, 17–32. [Google Scholar] [PubMed]
- Kotaka, M.; Johnson, C.; Lamb, H.K.; Hawkins, A.R.; Ren, J.; Stammers, D.K. Structural analysis of the recognition of the negative regulator NmrA and DNA by the zinc finger from the GATA-type transcription factor AreA. J. Mol. Biol. 2008, 381, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pan, Y.; Liu, G. Disruption of the nitrogen regulatory gene AcareA in Acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism. Fungal Genet. Biol. 2013, 61, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.Y.; Marletta, M.A.; Rine, J. Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum. Biochemistry 2008, 47, 7274–7283. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 656. [Google Scholar] [CrossRef] [PubMed]
- Pfannmüller, A.; Wagner, D.; Sieber, C.; Schönig, B.; Boeckstaens, M.; Marini, A.M.; Tudzynski, B. The general amino acid permease FfGap1 of Fusarium fujikuroi is sorted to the vacuole in a nitrogen-dependent, but Npr1 kinase-independent manner. PLoS ONE 2015, 10, e0125487. [Google Scholar] [CrossRef] [PubMed]
- Ravagnani, A.; Gorfinkiel, L.; Langdon, T.; Diallinas, G.; Adjadj, E.; Demais, S.; Gorton, D.; Arst, H.N., Jr.; Scazzocchio, C. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. EMBO J. 1997, 16, 3974–3986. [Google Scholar] [CrossRef] [PubMed]
- Muro-Pastor, M.I.; Strauss, J.; Ramón, A.; Scazzocchio, C. A paradoxical mutant GATA factor. Eukaryot. Cell 2004, 3, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Berger, H.; Pachlinger, R.; Morozov, I.; Goller, S.; Narendja, F.; Caddick, M.; Strauss, J. The GATA factor area regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol. Microbiol. 2006, 59, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Berger, H.; Basheer, A.; Böck, S.; Reyes-Dominguez, Y.; Dalik, T.; Altmann, F.; Strauss, J. dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol. Microbiol. 2008, 69, 1385–1398. [Google Scholar] [CrossRef] [PubMed]
- Haas, H.; Angermayr, K.; Zadra, I.; Stöffler, G. Overexpression of nreB, a new GATA factor-encoding gene of Penicillium chrysogenum, leads to repression of the nitrate assimilatory gene cluster. J. Biol. Chem. 1997, 272, 22576–22582. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.H.; Hynes, M.J.; Todd, R.B.; Davis, M.A. Deletion and overexpression of the Aspergillus nidulans GATA factor AreB reveals unexpected pleiotropy. Microbiology 2009, 155, 3868–3880. [Google Scholar] [CrossRef] [PubMed]
- Michielse, C.B.; Pfannmüller, A.; Macios, M.; Rengers, P.; Dzikowska, A.; Tudzynski, B. The interplay between the GATA transcription factors AreA, the global nitrogen regulator and AreB in Fusarium fujikuroi. Mol. Microbiol. 2014, 91, 472–493. [Google Scholar] [CrossRef] [PubMed]
- Pfannmüller, A.; Boysen, J.M.; Tudzynski, B. Nitrate Assimilation in Fusarium fujikuroi is controlled by multiple levels of regulation. Front. Microbiol. 2017, 8, 381. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.Q.; Heim, J.; Solomon, N.A.; Wolfe, S.; Demain, A.L. Repression of beta-lactam production in Cephalosporium acremonium by nitrogen sources. J. Antibiot. Tokyo 1984, 37, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Friedlin, E.; Marzluf, G.A. A reporter gene analysis of penicillin biosynthesis gene expression in Penicillium chrysogenum and its regulation by nitrogen and glucose catabolite repression. Appl. Environ. Microbiol. 1994, 60, 4432–4439. [Google Scholar] [PubMed]
- Haas, H.; Marzluf, G.A. NRE, the major nitrogen regulatory protein of Penicillium chrysogenum, binds specifically to elements in the intergenic promoter regions of nitrate assimilation and penicillin biosynthetic gene clusters. Curr. Genet. 1995, 28, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Menne, S.; Walz, M.; Kück, U. Expression studies with the bidirectional pcbAB-pcbC promoter region from Acremonium chrysogenum using reporter gene fusions. Appl. Microbiol. Biotechnol. 1994, 42, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Schjerling, P.; Holmberg, S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996, 24, 4599–4607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platt, A.; Reece, R.J. The yeast galactose genetic switch is mediated by the formation of a Gal4p–Gal80p–Gal3p complex. EMBO J. 1998, 14, 4086–4091. [Google Scholar] [CrossRef] [PubMed]
- Mamane, Y.; Hellauer, K.; Rochon, M.H.; Turcotte, B. A linker region of the yeast zinc cluster protein Leu3p specifies binding to everted repeat DNA. J. Biol. Chem. 1998, 273, 18556–18561. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Coleman, J.E. The DNA binding domain of Gal4 forms a binuclear metal ion complex. Biochemistry 1990, 29, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
- Marmostein, R.; Harrison, S.C. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 1994, 8, 2504–2512. [Google Scholar] [CrossRef]
- Anderson, S.F.; Steber, C.M.; Esposito, R.E.; Coleman, J.E. UME6, a negative regulator of meiosis in Saccharomyces cerevisiae, contains a C-terminal Zn2Cys6 binuclear cluster that binds the URS1 DNA sequence in a Zinc-dependent manner. Protein Sci. 1995, 4, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Marmostein, R.; Carey, M.; Ptashne, M.; Harrison, S.C. DNA recognition by Gal4: Structure of a protein-DNA complex. Nature 1992, 356, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Ehrlich, K.C. Genome-wide analysis of the Zn(II)2Cys6 zinc cluster-encoding gene family in Aspergillus flavus. Appl. Microbiol. Biotechnol. 2013, 97, 4289–4300. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, S.A.; Nekludova, L.; Pabo, C.O. DNA recognizion by Cys(2)His(2) zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 183–212. [Google Scholar] [CrossRef] [PubMed]
- Harbison, C.T.; Gordon, D.B.; Lee, T.I.; Rinaldi, N.J.; Macisaac, K.D.; Danford, T.W.; Hannett, N.M.; Tagne, J.B.; Reynolds, D.B.; Yoo, J.; et al. Transcriptional regulatory code of a eukaryotic genome. Nature 2004, 431, 99–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaestner, K.H.; Knochel, W.; Martinez, D.E. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000, 14, 142–146. [Google Scholar] [PubMed]
- Gajiwala, K.S.; Chen, H.; Cornille, F.; Roques, B.P.; Reith, W.; Mach, B.; Burley, S.K. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 2000, 403, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, E.K.; Kück, U. The fungal CPCR1 protein, which binds specifically to beta-lactam biosynthesis genes, is related to human regulatory factor X transcription factors. J. Biol. Chem. 2000, 275, 9348–9357. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, E.K.; Bunse, A.; Janus, D.; Hoff, B.; Friedlin, E.; Kürnsteiner, H.; Kück, U. Winged helix transcription factor CPCR1 is involved in regulation of beta-lactam biosynthesis in the fungus Acremonium chrysogenum. Eukaryot. Cell 2004, 3, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Hoff, B.; Schmitt, E.K.; Kück, U. CPCR1, but not its interacting transcription factor AcFKH1, controls fungal arthrospore formation in Acremonium chrysogenum. Mol. Microbiol. 2005, 56, 1220–1233. [Google Scholar] [CrossRef] [PubMed]
- Bugeja, H.E.; Hynes, M.J.; Andrianopoulos, A. The RFX Protein RfxA is an Essential Regulator of Growth and Morphogenesis in Penicillium marneffei. Eukaryot. Cell 2010, 9, 578–591. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Santos, R.; Martín, J.F.; Kosalková, K.; Prieto, C.; Ullán, R.V.; García-Estrada, C. The regulatory factor PcRFX1 controls the expression of the three genes of β-lactam biosynthesis in Penicillium chrysogenum. Fungal Genet. Biol. 2012, 49, 866–881. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, E.; Müller, D.; Knöchel, W. DNA recognition site analysis of Xenopus winged helix proteins. J. Mol. Biol. 1995, 248, 239–254. [Google Scholar] [CrossRef]
- Schmitt, E.K.; Hoff, B.; Kück, U. AcFKH1, a novel member of the forkhead family, associates with the RFX transcription factor CPCR1 in the cephalosporin C-producing fungus Acremonium chrysogenum. Gene 2004, 342, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Santos, R.; García-Estrada, C.; Kosalková, K.; Prieto, C.; Santamarta, I.; Martín, J.F. PcFKH1, a novel regulatory factor from the forkhead family, controls the biosynthesis of penicillin in Penicillium chrysogenum. Biochimie 2015, 115, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; Shimizu, K.; Keller, N.P. Genetics and biosynthesis of aflatoxins and sterigmatocystin. In The Mycota; Kempken, F., Bennett, J.W., Eds.; Springer-Verlag: Berlin, Germany, 2002; Volume 11, pp. 55–69. ISBN 3-540-42628-0. [Google Scholar]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Balajee, S.A.; Marr, K.A.; Andes, D.; Nielsen, K.F.; Frisvad, J.C.; Keller, N.P. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 2005, 4, 1574–1582. [Google Scholar] [CrossRef] [PubMed]
- Amaike, S.; Keller, N.P. Distinct Roles for VeA and LaeA in Development and Pathogenesis of Aspergillus flavus. Eukaryot. Cell 2009, 7, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.Y.; Hou, Y.; Chen, Y.; Oh, J.H.; Lee, I.; Bugni, T.S.; Keller, N.P. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl. Environ. Microbiol. 2012, 78, 4117–4125. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F. Key role of LaeA and velvet complex proteins on expression of β-lactam and PR-toxin genes in Penicillium chrysogenum: cross-talk regulation of secondary metabolite pathways. J. Ind. Microbiol. Biotechnol. 2017, 44, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.C.; Zhang, X.; Trievel, R.C.; Cheng, X. The SET-domain protein superfamily: Protein lysine methyltransferases. Genome Biol. 2005, 6, 227. [Google Scholar] [CrossRef] [PubMed]
- Kosalková, K.; García-Estrada, C.; Ullán, R.V.; Godio, R.P.; Feltrer, R.; Teijeira, F.; Mauriz, E.; Martín, J.F. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 2009, 91, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Keller, N.P. Secondary metabolism in fungi: Does chromosomal location matter? Curr. Opin. Microbiol. 2010, 13, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Hoff, B.; Kamerewerd, J.; Sigl, C.; Mitterbauer, R.; Zadra, I.; Kürnsteiner, H.; Kück, U. Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum, Eukaryot. Cell 2010, 9, 1236–1250. [Google Scholar] [CrossRef]
- Bok, J.W.; Chiang, Y.M.; Szewczyk, E.; Reyes-Dominguez, Y.; Davidson, A.D.; Sanchez, J.F.; Lo, H.C.; Watanabe, K.; Strauss, J.; Oakley, B.R.; et al. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009, 5, 462–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strauss, J.; Reyes-Dominguez, Y. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol. 2011, 48, 62–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Dominguez, Y.; Bok, J.W.; Berger, H.; Shwab, E.K.; Basheer, A.; Gallmetzer, A.; Scazzocchio, C.; Keller, N.; Strauss, J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 2010, 76, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Patananan, A.N.; Palmer, J.M.; Garvey, G.S.; Keller, N.P.; Clarke, S.G. A novel automethylation reaction in the Aspergillus nidulans LaeA protein generates S-methylmethionine. J. Biol. Chem. 2013, 288, 14032–14045. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.; García-Estrada, C.; Kosalková, K.; Ullán, R.V.; Albillos, S.M.; Martín, J.F. The inducers 1,3-diaminopropane and spermidine produce a drastic increase in the expression of the penicillin biosynthetic genes for prolonged time, mediated by the laeA regulator. Fungal Genet. Biol. 2012, 49, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Bayram, O.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Kopke, K.; Hoff, B.; Bloemendal, S.; Katschorowski, A.; Kamerewerd, J.; Kuck, U. Members of the Penicillium chrysogenum velvet complex play functionally opposing roles in the regulation of penicillin biosynthesis and conidiation. Eukaryot. Cell 2013, 12, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Brown, D.W.; Kleigrewe, K.; Bok, J.W.; Keller, N.P.; Humpf, H.U.; Tudzynski, B. FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol. Microbiol. 2010, 77, 972–994. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Oide, S.; Zhang, N.; Choi, M.Y.; Turgeon, B.G. ChLae1 and ChVel1 regulate T-toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus. PLoS Pathog. 2012, 8, e1002542. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Nam, T.Y.; Han, K.H.; Kim, S.C.; Yu, J.H. VelC positively controls sexual development in Aspergillus nidulans. PLoS ONE 2014, 9, e89883. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, J.; Eichhorn, H.; Friedlin, E.; Kurnsteiner, H.; Kuck, U. A homologue of the Aspergillus velvet gene regulates both cephalosporin C biosynthesis and hyphal fragmentation in Acremonium chrysogenum. Appl. Environ. Microbiol. 2007, 73, 3412–3422. [Google Scholar] [CrossRef] [PubMed]
- Terfehr, D.; Dahlmann, T.A.; Kück, U. Transcriptome analysis of the two unrelated fungal β-lactam producers Acremonium chrysogenum and Penicillium chrysogenum: Velvet-regulated genes are major targets during conventional strain improvement programs. BMC Genom. 2017, 18, 272. [Google Scholar] [CrossRef] [PubMed]







| Fungal Species | Transcription Factor | Process/Function | Reference |
|---|---|---|---|
| Ascomycetes | |||
| Acremonium chrysogenum | Cre1 | Glucose catabolite regulation | [19,20] |
| Aspergillus nidulans | PacC | pH regulation | [21] |
| Aspergillus nidulans | AslA | Control of the K(+) stress-inducible expression of the genes encoding the ion pumps | [22] |
| Aspergillus nidulans | SltA | Control of morphogenesis and sterigmatocystin biosynthesis | [23] |
| Aspergillus nidulans | FlbC | Control of conidial development and germination, and brlA and vosA expression | [24] |
| Aspergillus nidulans | NsdC | Control of fruiting body formation | [25] |
| Aspergillus nidulans | MtfA | Regulation of secondary metabolism and differentiation | [26] |
| Aspergillus nidulans | BrlA | Regulation of differentiation | [27] |
| Aspergillus nidulans, Penicillium chrysogenum | CreA | Glucose catabolite regulation | [28,29] |
| Aspergillus glaucus | Agseb1 | Control of sensitivity to hyperosmotic stress. Hyphae branching and aspergiolide A biosynthesis | [30] |
| Botrytis cinerea | BcYOH1 | Control of botrydial, phytotoxin and other secondary metabolites gene clusters | [31] |
| Colletotrichum gloeosporioides | CrzA | Control of morphogenetic processes and pathogenicity | [32] |
| Fusarium verticillioides | Ada1 | Control of asexual development | [33] |
| Fusarium verticillioides | Sda1 | Control of polyol metabolism and fumonisin biosynthesis | [34] |
| Magnaporthe oryzae | Znf1 | Control of pathogenicity | [35] |
| Magnaporthe oryzae | Flb3p | Required for normal aerial mycelium formation | [36] |
| Magnaporthe oryzae | Cos1 | Control of conidiophores development | [37] |
| Saccharomyces cerevisiae | MIG1 | Glucose catabolite regulation | [38] |
| Trichoderma atroviride | Blu7 | Control of light-regulated genes, growth and conidiation | [39] |
| Verticillium dahliae | Czf | Control of fungal growth, development, various stress responses, and virulence | [40] |
| Basidiomycetes | |||
| Agaricus bisporus | C2H2 | Control of timing for mushroom formation | [41] |
| Cryptococcus neoformans | Ste12αp | Regulation of several genes associated with virulence | [42] |
| Ganoderma lucidum | GlPacC | Control of response to ambient pH | [43] |
| Schizophyllum commune | C2H2 | Regulation of primordium development | [44] |
| Ustilago maydis | Mzr1 | Transcriptional activator during host colonization | [45] |
| Ustilago maydis | Rua1 | Regulation of biosurfactant ustilagic acid biosynthesis | [46] |
| Fungal Species | Transcription Factor | Process/Function | Reference |
|---|---|---|---|
| Ascomycetes | |||
| Aspergillus nidulans | AclR | Activation of genes for ethanol oxidation | [47,48] |
| Aspergillus nidulans | ArcA | Arginine catabolic pathway | [49] |
| Aspergillus nidulans | NirA | Activation of nitrate assimilation | [50,51] |
| Aspergillus nidulans | PrnA | Activation of proline utilization | [52,53] |
| Aspergillus nidulans | QutA | Regulation of genes for quinic acid utilization | [54] |
| Aspergillus nidulans | QutH | Possible role in the regulation of protocatechuic acid utilization | [55] |
| Aspergillus nidulans | TamA | Nitrogen regulation | [56] |
| Aspergillus nidulans | Uay | Activation of purine transport and utilization | [57,58] |
| Aspergillus nidulans, Aspergillus oryzae, Aspergillus niger | AmyR | Activation of amylolytic gene expression | [59,60] |
| Aspergillus nidulans, Neurospora crassa | FacB | Activation of acetate regulatory genes | [61,62] |
| Aspergillus oryzae, Aspergillus niger, Fusarium oxysporum | XlnR | Regulation of xylanolytic genes expression | [63] |
| Aspergillus fumigatus | GliZ | Regulation of gliotoxin | [64] |
| Bipolaris oryzae | Bmr1 | Regulation of melanin biosynthesis | [65] |
| Clonostachys rogersoniana | VerZ | Regulation of verticillin biosynthesis | [66] |
| Colletotricum lindemuthianum | CltA1 | Involved in the regulation of biotrophy/nectotrophy switch | [67] |
| Colletotricum lagenarium | CmR1 | Regulation of melanin biosynthesis | [68] |
| Cercospora nicotianae | CRG1 | Involved in cercosporin resistance | [69] |
| Fusarium verticillioides | Zrf1 | Regulation of fumonisin biosynthesis and sugars transport | [70] |
| Fusarium oxysporum | Fow2 | Control of pathogenicity | [71] |
| Fusarium graminearum and F. verticillioides | FgArt1 | Regulation of tricotecenos and fumonisin. | [72] |
| Fusarium graminearum | Ebr1 | Involved in growth and virulence | [73] |
| Leptosphaeria maculans | SirZ | Regulation of sirodesmin biosynthesis | [74] |
| Magnaporthe grisea | Pig1 | Regulation of melanin biosynthesis | [68] |
| Nectria haematococca | Ctf1 a and b | Activation of cutinases | [75] |
| Neurospora crassa | Fl | Required for conidiophore morphogenesis | [76] |
| Neurospora crassa | Nit4 | Activaion of nitrate assimilation | [77] |
| Penicillium citrinum | MlcR | Involved in compactin biosynthesis | [78] |
| Penicillium roqueforti | Pcz1 | Regulation of cell development | [79] |
| Parastagonospora nodorum | PnPf2 | Involved in host specific virulence | [80] |
| Saccharomyces cerevisiae | ArgRII | Control of arginine metabolism | [81] |
| Talaromyces marneffei | FacB | Regulation of the glyoxalate cycle | [82] |
| Trichoderma reesei | AceII | Activation of cellulases and xylanases gene expression | [83] |
| Sordaria macrospora | Pro1 | Involved in the development of fruiting bodies | [84] |
| Basidiomycetes | |||
| Schizophyllum commune | Fst3 | Regulation of mushroom development | [85] |
| Schizophyllum commune | Fst4 | Regulation of mushroom formation | [85] |
| Ustilago maydis | Ton1 | Involved in inducing the expression of rrm4 (an RNA-binding protein involved in cell polarity and also required for full pathogenicity) | [86] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Estrada, C.; Domínguez-Santos, R.; Kosalková, K.; Martín, J.-F. Transcription Factors Controlling Primary and Secondary Metabolism in Filamentous Fungi: The β-Lactam Paradigm. Fermentation 2018, 4, 47. https://doi.org/10.3390/fermentation4020047
García-Estrada C, Domínguez-Santos R, Kosalková K, Martín J-F. Transcription Factors Controlling Primary and Secondary Metabolism in Filamentous Fungi: The β-Lactam Paradigm. Fermentation. 2018; 4(2):47. https://doi.org/10.3390/fermentation4020047
Chicago/Turabian StyleGarcía-Estrada, Carlos, Rebeca Domínguez-Santos, Katarina Kosalková, and Juan-Francisco Martín. 2018. "Transcription Factors Controlling Primary and Secondary Metabolism in Filamentous Fungi: The β-Lactam Paradigm" Fermentation 4, no. 2: 47. https://doi.org/10.3390/fermentation4020047
APA StyleGarcía-Estrada, C., Domínguez-Santos, R., Kosalková, K., & Martín, J. -F. (2018). Transcription Factors Controlling Primary and Secondary Metabolism in Filamentous Fungi: The β-Lactam Paradigm. Fermentation, 4(2), 47. https://doi.org/10.3390/fermentation4020047