Non-Coding RNAs, COVID-19, and Long-COVID
Share This Topical Collection
Editors
 Prof. Dr. Gaetano Santulli
Prof. Dr. Gaetano Santulli
 Prof. Dr. Gaetano Santulli
Prof. Dr. Gaetano Santulli
E-Mail
Website
Collection Editor
1. Department of Medicine, Fleischer Institute for Diabetes and Metabolism (FIDAM), Einstein-Mount Sinai Diabetes Research Center (ES-DRC), Albert Einstein College of Medicine, New York, NY 10461, USA
2. Department of Advanced Biomedical Science, “Federico II” University, International Translational Research and Medical Education Consortium (ITME), 80131 Naples, Italy
3. Department of Molecular Pharmacology, Albert Einstein College of Medicine, New York, NY 10461, USA
Interests: cardiology; hypertension; restenosis; heart failure; myocardial infarction; endothelial dysfunction; mitochondria; diabetes; microRNAs; insulin resistance; atherosclerosis; thrombosis; cardiac hypertrophy; pancreatic beta cell function; insulin release
Special Issues, Collections and Topics in MDPI journals
 Dr. Jessica Gambardella
Dr. Jessica Gambardella
 Dr. Jessica Gambardella
Dr. Jessica Gambardella
E-Mail
Website
Collection Editor
Department of Molecular Pharmacology, Albert Einstein College of Medicine - Montefiore University Hospital, 1300 Morris Park Avenue, New York, NY 10461, USA
Interests: molecular biology; calcium; endothelium; mitochondria; heart diseases; microRNAs
Topical Collection Information
Dear Colleagues,
Recent evidence has shown that non-coding RNAs could be implemented in the pathophysiology of the systemic manifestations of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus (SARS-CoV-2), and most importantly, in long-COVID.
On these grounds, this Topical Collection of Non-Coding RNA has the main objective to gather high-quality research papers that have a solid basic research setting and/or translational potential on this topic. Therefore, studies exploring the mechanistic role of non-coding RNAs in the pathophysiology of COVID-19 and its complications, including long-COVID are especially welcome in this Topical Collection. We are interested in both clinical and pre-clinical investigations testing novel hypotheses that can advance the field. Novel therapeutic hypotheses and medicinal chemistry endeavors toward novel drug development are also encouraged.
In this Topical Collection, we are making a call to action to stimulate researchers to submit their invaluable studies on non-coding RNAs, COVID-19, and long-COVID. Original investigations, as well as review articles, are welcome. In summary, this Topical Collection aims to underscore the key importance of non-coding RNAs in COVID-19 and long-COVID.
Prof. Dr. Gaetano Santulli
Dr. Jessica Gambardella
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Non-Coding RNA is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 1800 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- COVID-19
- long-COVID
- RNA biology
- microRNA
- lncRNA
- circular RNA
- SARS-CoV-2
- exosomes
- virus
- vaccine
- epigenetics
- biomarkers
- epitranscriptomics
- bioinformatics
Published Papers (6 papers)
Open AccessArticle
Circulating miRNAs in the Plasma of Post-COVID-19 Patients with Typical Recovery and Those with Long-COVID Symptoms: Regulation of Immune Response-Associated Pathways
by
Anna M. Timofeeva, Artem O. Nikitin and Georgy A. Nevinsky
Viewed by 1775
Abstract
Following the acute phase of SARS-CoV-2 infection, certain individuals experience persistent symptoms referred to as long COVID. This study analyzed the patients categorized into three distinct groups: (1) individuals presenting rheumatological symptoms associated with long COVID, (2) patients who have successfully recovered from
[...] Read more.
Following the acute phase of SARS-CoV-2 infection, certain individuals experience persistent symptoms referred to as long COVID. This study analyzed the patients categorized into three distinct groups: (1) individuals presenting rheumatological symptoms associated with long COVID, (2) patients who have successfully recovered from COVID-19, and (3) donors who have never contracted COVID-19. A notable decline in the expression of miR-200c-3p, miR-766-3p, and miR-142-3p was identified among patients exhibiting rheumatological symptoms of long COVID. The highest concentration of miR-142-3p was found in healthy donors. One potential way to reduce miRNA concentrations is through antibody-mediated hydrolysis. Not only can antibodies possessing RNA-hydrolyzing activity recognize the miRNA substrate specifically, but they also catalyze its hydrolysis. The analysis of the catalytic activity of plasma antibodies revealed that antibodies from patients with long COVID demonstrated lower hydrolysis activity against five fluorescently labeled oligonucleotide sequences corresponding to the Flu-miR-146b-5p, Flu-miR-766-3p, Flu-miR-4742-3p, and Flu-miR-142-3p miRNAs and increased activity against the Flu-miR-378a-3p miRNA compared to other patient groups. The changes in miRNA concentrations and antibody-mediated hydrolysis of miRNAs are assumed to have a complex regulatory mechanism that is linked to gene pathways associated with the immune system. We demonstrate that all six miRNAs under analysis are associated with a large number of signaling pathways associated with immune response-associated pathways.
Full article
►▼
Show Figures
Open AccessCommunication
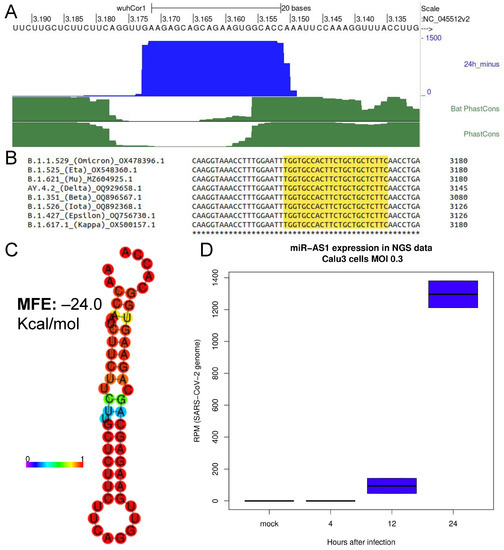
A microRNA Arising from the Negative Strand of SARS-CoV-2 Genome Targets FOS to Reduce AP-1 Activity
by
Francesco Greco, Elisa Lorefice, Claudia Carissimi, Ilaria Laudadio, Fabiola Ciccosanti, Martina Di Rienzo, Francesca Colavita, Silvia Meschi, Fabrizio Maggi, Gian Maria Fimia and Valerio Fulci
Cited by 2 | Viewed by 2363
Abstract
Virus-encoded microRNAs were first reported in the Epstein–Barr virus in 2004. Subsequently, a few hundred viral miRNAs have been identified, mainly in DNA viruses belonging to the
herpesviridae family. To date, only 30 viral miRNAs encoded by RNA viruses are reported by miRBase.
[...] Read more.
Virus-encoded microRNAs were first reported in the Epstein–Barr virus in 2004. Subsequently, a few hundred viral miRNAs have been identified, mainly in DNA viruses belonging to the
herpesviridae family. To date, only 30 viral miRNAs encoded by RNA viruses are reported by miRBase. Since the outbreak of the SARS-CoV-2 pandemic, several studies have predicted and, in some cases, experimentally validated miRNAs originating from the positive strand of the SARS-CoV-2 genome. By integrating NGS data analysis and qRT-PCR approaches, we found that SARS-CoV-2 also encodes for a viral miRNA arising from the minus (antisense) strand of the viral genome, in the region encoding for ORF1ab, herein referred to as SARS-CoV-2-miR-AS1. Our data show that the expression of this microRNA increases in a time course analysis of SARS-CoV-2 infected cells. Furthermore, enoxacin treatment enhances the accumulation of the mature SARS-CoV-2-miR-AS1 in SARS-CoV-2 infected cells, arguing for a Dicer-dependent processing of this small RNA. In silico analysis suggests that SARS-CoV-2-miR-AS1 targets a set of genes which are translationally repressed during SARS-CoV-2 infection. We experimentally validated that SARS-CoV-2-miR-AS1 targets FOS, thus repressing the AP-1 transcription factor activity in human cells.
Full article
►▼
Show Figures
Open AccessArticle
Co-Regulation of Protein Coding Genes by Transcription Factor and Long Non-Coding RNA in SARS-CoV-2 Infected Cells: An In Silico Analysis
by
Chinmay Saha, Sayantan Laha, Raghunath Chatterjee and Nitai P. Bhattacharyya
Cited by 5 | Viewed by 4550
Abstract
Altered expression of protein coding gene (PCG) and long non-coding RNA (lncRNA) have been identified in SARS-CoV-2 infected cells and tissues from COVID-19 patients. The functional role and mechanism (s) of transcriptional regulation of deregulated genes in COVID-19 remain largely unknown. In the
[...] Read more.
Altered expression of protein coding gene (PCG) and long non-coding RNA (lncRNA) have been identified in SARS-CoV-2 infected cells and tissues from COVID-19 patients. The functional role and mechanism (s) of transcriptional regulation of deregulated genes in COVID-19 remain largely unknown. In the present communication, reanalyzing publicly available gene expression data, we observed that 66 lncRNA and 5491 PCG were deregulated in more than one experimental condition. Combining our earlier published results and using different publicly available resources, it was observed that 72 deregulated lncRNA interacted with 3228 genes/proteins. Many targets of deregulated lncRNA could also interact with SARS-CoV-2 coded proteins, modulated by IFN treatment and identified in CRISPR screening to modulate SARS-CoV-2 infection. The majority of the deregulated lncRNA and PCG were targets of at least one of the transcription factors (TFs), interferon responsive factors (IRFs), signal transducer, and activator of transcription (STATs), NFκB, MYC, and RELA/p65. Deregulated 1069 PCG was joint targets of lncRNA and TF. These joint targets are significantly enriched with pathways relevant for SARS-CoV-2 infection indicating that joint regulation of PCG could be one of the mechanisms for deregulation. Over all this manuscript showed possible involvement of lncRNA and mechanisms of deregulation of PCG in the pathogenesis of COVID-19.
Full article
►▼
Show Figures
Open AccessPerspective
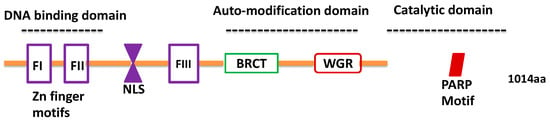
Therapeutic Significance of microRNA-Mediated Regulation of PARP-1 in SARS-CoV-2 Infection
by
Sabyasachi Dash, Chandravanu Dash and Jui Pandhare
Cited by 16 | Viewed by 4664
Abstract
The COVID-19 pandemic caused by the novel coronavirus SARS-CoV-2 (2019-nCoV) has devastated global healthcare and economies. Despite the stabilization of infectivity rates in some developed nations, several countries are still under the grip of the pathogenic viral mutants that are causing a significant
[...] Read more.
The COVID-19 pandemic caused by the novel coronavirus SARS-CoV-2 (2019-nCoV) has devastated global healthcare and economies. Despite the stabilization of infectivity rates in some developed nations, several countries are still under the grip of the pathogenic viral mutants that are causing a significant increase in infections and hospitalization. Given this urgency, targeting of key host factors regulating SARS-CoV-2 life cycle is postulated as a novel strategy to counter the virus and its associated pathological outcomes. In this regard, Poly (ADP)-ribose polymerase-1 (PARP-1) is being increasingly recognized as a possible target. PARP-1 is well studied in human diseases such as cancer, central nervous system (CNS) disorders and pathology of RNA viruses. Emerging evidence indicates that regulation of PARP-1 by non-coding RNAs such as microRNAs is integral to cell survival, redox balance, DNA damage response, energy homeostasis, and several other cellular processes. In this short perspective, we summarize the recent findings on the microRNA/PARP-1 axis and its therapeutic potential for COVID-19 pathologies.
Full article
►▼
Show Figures
Open AccessReview
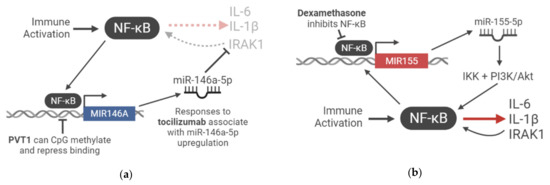
Non-Coding RNAs in COVID-19: Emerging Insights and Current Questions
by
Tobias Plowman and Dimitris Lagos
Cited by 25 | Viewed by 5843
Abstract
The highly infectious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged as the causative agent of coronavirus disease 2019 (COVID-19) in late 2019, igniting an unprecedented pandemic. A mechanistic picture characterising the acute immunopathological disease in severe COVID-19 is developing. Non-coding RNAs (ncRNAs)
[...] Read more.
The highly infectious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged as the causative agent of coronavirus disease 2019 (COVID-19) in late 2019, igniting an unprecedented pandemic. A mechanistic picture characterising the acute immunopathological disease in severe COVID-19 is developing. Non-coding RNAs (ncRNAs) constitute the transcribed but un-translated portion of the genome and, until recent decades, have been undiscovered or overlooked. A growing body of research continues to demonstrate their interconnected involvement in the immune response to SARS-CoV-2 and COVID-19 development by regulating several of its pathological hallmarks: cytokine storm syndrome, haemostatic alterations, immune cell recruitment, and vascular dysregulation. There is also keen interest in exploring the possibility of host–virus RNA–RNA and RNA–RBP interactions. Here, we discuss and evaluate evidence demonstrating the involvement of short and long ncRNAs in COVID-19 and use this information to propose hypotheses for future mechanistic and clinical studies.
Full article
►▼
Show Figures
Open AccessCommentary
microRNA Heterogeneity, Innate-Immune Defense and the Efficacy of SARS-CoV-2 Infection—A Commentary
by
Walter J. Lukiw
Cited by 14 | Viewed by 3916
Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a member of the genus
Betacoronavirus in the family
Coronaviridae, possesses an unusually large single-stranded viral RNA (ssvRNA) genome of about ~29,811 nucleotides (nt) that causes severe and acute respiratory distress and a highly lethal viral
[...] Read more.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a member of the genus
Betacoronavirus in the family
Coronaviridae, possesses an unusually large single-stranded viral RNA (ssvRNA) genome of about ~29,811 nucleotides (nt) that causes severe and acute respiratory distress and a highly lethal viral pneumonia known as COVID-19. COVID-19 also presents with multiple ancillary systemic diseases and often involves cardiovascular, inflammatory, and/or neurological complications. Pathological viral genomes consisting of ssvRNA, like cellular messenger RNA (mRNA), are susceptible to attack, destruction, neutralization, and/or modulation by naturally occurring small non-coding RNAs (sncRNAs) within the host cell, some of which are known as microRNAs (miRNAs). This paper proposes that the actions of the 2650 known human miRNAs and other sncRNAs form the basis for an under-recognized and unappreciated innate-immune regulator of ssvRNA viral genome activities and have implications for the efficiency of SARS-CoV-2 invasion, infection, and replication. Recent research indicates that both miRNA and mRNA abundance, speciation, and complexity varies widely amongst human individuals, and this may:
(i) In part explain the variability in the innate-immune immunological and pathophysiological response of different human individuals to the initiation and progression of SARS-CoV-2 infection in multiple tissue types; and
(ii) further support our understanding of human biochemical and genetic individuality and the variable resistance of individuals to ssvRNA-mediated viral infection and disease. This commentary will briefly address current findings and concepts in this fascinating research area of non-coding RNA and innate-immunity with special reference to natural host miRNAs, SARS-CoV-2, and the current COVID-19 pandemic.
Full article
Planned Papers
The below list represents only planned manuscripts. Some of these
manuscripts have not been received by the Editorial Office yet. Papers
submitted to MDPI journals are subject to peer-review.
Title: Non-coding RNAs in COVID-19: Emerging insights and current questions
Authors: Dimitris Lagos; Toby Plowman
Affiliation: Hull York Medical School
Abstract: To be determined