Novel Bioactive Paulomycin Derivatives Produced by Streptomyces albus J1074
Abstract
:1. Introduction
2. Results
2.1. Identification of Compounds
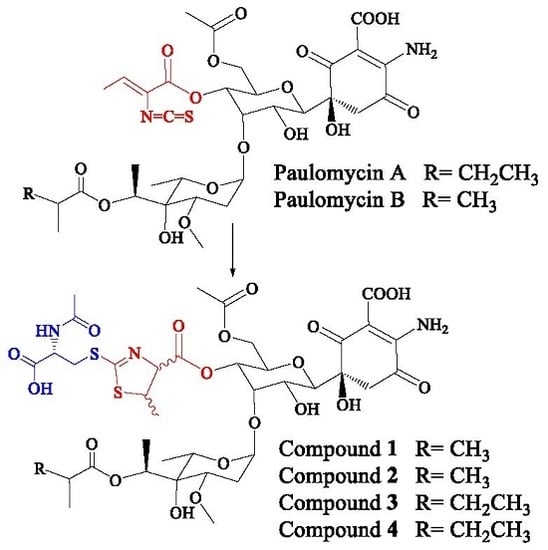
2.2. Structural Characterization of Novel Paulomycins
2.3. Biological Activity of Novel Paulomycins
2.4. Stability of Novel Paulomycins in Culture
3. Discussion
4. Materials and Methods
4.1. Microorganisms and Culture Media
4.2. Isolation and Structural Characterization of Compounds
4.3. Biological Activity
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chater, K.F.; Wilde, L.C. Restriction of a bacteriophage of Streptomyces albus G involving endonuclease Sall. J. Bacteriol. 1976, 128, 644–650. [Google Scholar] [PubMed]
- Baltz, R.H. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J. Ind. Microbiol. Biotechnol. 2010, 37, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Zaburannyi, N.; Rabyk, M.; Ostash, B.; Fedorenko, V.; Luzhetskyy, A. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Tokovenko, B.; Brötz, E.; Rückert, C.; Kalinowski, J.; Luzhetskyy, A. Genome rearrangements of Streptomyces albus J1074 lead to the carotenoid gene cluster activation. Appl. Microbiol. Biotechnol. 2014, 98, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; García, I.; González, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sánchez-Hidalgo, M.; Braña, A.F.; Méndez, C.; Salas, J.A. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J. Am. Chem. Soc. 2017. [Google Scholar] [CrossRef] [PubMed]
- Majer, J.; Chater, K.F. Streptomyces albus G produces an antibiotic complex identical to paulomycins A and B. J. Gen. Microbiol. 1987, 133, 2503–2507. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Rodríguez, M.; Braña, A.F.; Méndez, C.; Salas, J.A.; Olano, C. New insights into paulomycin biosynthesis pathway in Streptomyces albus J1074 and generation of novel derivatives by combinatorial biosynthesis. Microb. Cell Fact. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Wiley, P.F. A new antibiotic U-43120 (NSC-163500). J. Antibiot. 1976, 29, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Argoudelis, A.D.; Brinkley, T.A.; Brodasky, T.F.; Buege, J.A.; Meyer, H.F.; Mizsak, S.A. Paulomycins A and B. Isolation and characterization. J. Antibiot. 1982, 35, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Argoudelis, A.D.; Baczynskyj, L.; Haak, W.J.; Knoll, W.M.; Mizsak, S.A.; Shilliday, F.B. New paulomycins produced by Streptomyces paulus. J. Antibiot. 1988, 41, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Argoudelis, A.D.; Baczynskyj, L.; Mizsak, S.A.; Shilliday, F.B. O-demethylpaulomycins A and B, U-77,802 and U-77,803, paulomenols A and B, new metabolites produced by Streptomyces paulus. J. Antibiot. 1988, 41, 1316–1330. [Google Scholar] [CrossRef] [PubMed]
- Argoudelis, A.D.; Baczynskyj, L.; Buege, J.A.; Marshall, V.P.; Mizsak, S.A.; Wiley, P.F. Paulomycin-related antibiotics: Paldimycins and antibiotics 273a2. Isolation and characterization. J. Antibiot. 1987, 40, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Argoudelis, A.D.; Baczynskyj, L.; Mizsak, S.A.; Shilliday, F.B.; Spinelli, P.A.; DeZwaan, J. Paldimycins A and B and antibiotics 273a2α and 273a2β. Synthesis and characterization. J. Antibiot. 1987, 40, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Eliopoulos, G.M.; Reiszner, E.; Moellering, R.C., Jr. In vitro evaluation of the new paulomycin antibiotic paldimycin. Eur. J. Clin. Microbiol. 1987, 6, 306–308. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; Méndez, C.; Salas, J.A. Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Nat. Prod. Rep. 2010, 27, 571–616. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; Méndez, C.; Salas, J.A. Molecular insights on the biosynthesis of antitumour compounds by actinomycetes. Microb. Biotechnol. 2011, 4, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Zarins-Tutt, J.S.; Barberi, T.T.; Gao, H.; Mearns-Spragg, A.; Zhang, L.; Newman, D.J.; Goss, R.J. Prospecting for new bacterial metabolites: A glossary of approaches for inducing, activating andupregulating the biosynthesis of bacterial cryptic or silent natural products. Nat. Prod. Rep. 2016, 33, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Wiley, P.F.; Mizsak, S.A.; Baczynskyj, L.; Argoudelis, A.D. The structure of paulomycin. J. Antibiot. 1984, 37, 1273–1275. [Google Scholar] [CrossRef] [PubMed]
- Wiley, P.F.; Mizsak, S.A.; Baczynskyj, L.; Argoudelis, A.D.; Duchamp, D.J.; Watt, W. The structure and chemistry of paulomycin. J. Org. Chem. 1986, 51, 2493–2499. [Google Scholar] [CrossRef]
- Sarmiento-Vizcaíno, A.; Braña, A.F.; Pérez-Victoria, I.; Martín, J.; de Pedro, N.; Cruz, M.; Díaz, C.; Vicente, F.; Acuña, J.L.; Reyes, F.; et al. Paulomycin G, a new natural product with cytotoxic activity against tumor cell lines produced by deep-sea sediment derived Micromonospora matsumotoense M-412 from the Avilés canyon in the Cantabrian sea. Mar. Drugs. 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Camery, J.R.; Hong, S.T.; Gould, S.J. Seongomycin: A new sulfur-containing benzo[b]fluorene derived from genes clustered with those for kanamycin biosynthesis. Tetrahedron Lett. 1997, 38, 3139–3142. [Google Scholar]
- Gilpin, M.L.; Fulston, M.; Payne, D.; Cramp, R.; Hood, I. Isolation and structure determination of two novel phenazines from a Streptomyces with inhibitory activity against metallo-enzymes, including metallo-β-lactamase. J. Antibiot. 1995, 48, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Hooper, A.M.; Rickards, R.W. 3-Amino-5-hydroxybenzoic acid in antibiotic biosynthesis. XI. Biological origins and semisynthesis of thionaphthomycins, and the structures of naphthomycins I and J. J. Antibiot. 1998, 51, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Omura, S.; Fujimoto, T.; Otoguro, K.; Matsuzaki, K.; Moriguchi, R.; Tanaka, H.; Sasaki, Y. Lactacystin, a novel microbial metabolite, induces neuritogenesis of neuroblastoma cells. J. Antibiot. 1991, 44, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M.; Bitzer, J.; Mayer-Bartschmid, A.; Müller, H.; Benet-Buchholz, J.; Gantner, F.; Tichy, H.V.; Reinemer, P.; Bacon, K.B. Cinnabaramides A–G: Analogues of lactacystin and salinosporamide from a terrestrial streptomycete. J. Nat. Prod. 2007, 70, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Schulz, D.; Nachtigall, J.; Riedlinger, J.; Schneider, K.; Poralla, K.; Imhoff, J.F.; Beil, W.; Nicholson, G.; Fiedler, H.P.; Süssmuth, R.D. Piceamycin and its N-acetylcysteine adduct is produced by Streptomyces sp. GB 4-2. J. Antibiot. 2009, 62, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Escribano, J.P.; Song, L.; Fox, D.J.; Yeo, V.; Bibb, M.J.; Challis, G.L. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 2012, 3, 2716–2720. [Google Scholar] [CrossRef]
- Woo, C.M.; Gholap, S.L.; Herzon, S.B. Insights into lomaiviticin biosynthesis. Isolation and structure elucidation of (−)-homoseongomycin. J. Nat. Prod. 2013, 76, 1238–1241. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Shin-Ya, K.; Nagamitsu, T.; Tomoda, H. Biosynthesis of mercapturic acid derivative of the labdane-type diterpene, cyslabdan that potentiates imipenem activity against methicillin-resistant Staphylococcus aureus: Cyslabdan is generated by mycothiol-mediated xenobiotic detoxification. J. Ind. Microbiol. Biotechnol. 2016, 43, 325–342. [Google Scholar] [CrossRef] [PubMed]
- Suhui, Y.; Molloy, B.; Braña, A.F.; Zabala, D.; Olano, C.; Cortés, J.; Moris, J.F.; Salas, J.A.; Méndez, C. Identification by genome mining of a type I polyketide gene cluster from Streptomyces argillaceus involved in the biosynthesis of pyridine and piperidine alkaloids argimycins P. Front. Microbiol. 2016, 15. [Google Scholar] [CrossRef]
- Fukumoto, A.; Kim, Y.P.; Hanaki, H.; Shiomi, K.; Tomoda, H.; Omura, S. Cyslabdan, a new potentiator of imipenem activity against methicillin-resistant Staphylococcus aureus, produced by Streptomyces sp. K04-0144 II. Biological activities. J. Antibiot. 2008, 61, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Fernández, E.; Weissbach, U.; Sánchez Reillo, C.; Braña, A.F.; Méndez, C.; Rohr, J.; Salas, J.A. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J. Bacteriol. 1998, 180, 4929–4937. [Google Scholar] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; Current Protocols: Brooklyn, NY, USA, 1994. [Google Scholar]
- Braña, A.F.; Rodríguez, M.; Pahari, P.; Rohr, J.; García, L.A.; Blanco, G. Activation and silencing of secondary metabolites in Streptomyces albus and Streptomyces lividans after transformation with cosmids containing the thienamycin gene cluster from Streptomyces cattleya. Arch. Microbiol. 2014, 196, 345–355. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–4 are available from the authors. |







| Position | 𝛅(13C) | 𝛅(1H), (Mult, J in Hz) | Position | 𝛅(13C) | 𝛅(1H), (Mult, J in Hz) |
|---|---|---|---|---|---|
| 1 | n.d. | - | 1′ | 98.5 | 4.89 (br d, 3.5) |
| 2 | 99.9 | - | 2′ | 30.7 | 2.04 (m) 1.77 (td, 12.3, 3.6) |
| 3 | 159.9 | - | 3′ | 75.0 | 3.43 (m) |
| 4 | 189.2 | - | 3′α | 57.1 | 3.24 (s) |
| 5 | 48.9 | 3.27 (m) 3.00 (d, 16.2) | 4′ | 73.9 | - |
| 6 | 78.2 | - | 5′ | 67.5 | 4.41 (m) |
| 7 | 198.7 | - | 6′ | 16.4 | 1.10 (m) |
| 8 | 77.7 | 3.64 (d, 10.0) | 7′ | 69.9 | 5.27 (quart., 6.8) |
| 9 | 69.2 | 3.48 (m) | 8′ | 16.5 | 1.18 (d, 6.7) |
| 10 | 74.5 | 4.08 (br t) | 9′ | 176.5 | - |
| 11 | 70.7 | 4.66 (dd, 10.2, 1.8) | 10′ | 34.3 | 2.55 (m) |
| 12 | 71.8 | 4.01 (dt, 9.9, 3.4) | 11′ | 19.6 | 1.08 (m) |
| 13 | 62.9 | 3.77 (m) | 12′ | 19.8 | 1.13 (d, 7.0) |
| 14 | 171.0 | - | 1′′ | 168.5 | - |
| 15 | 21.2 | 1.96 (s) | 2′′ | 80.3 | 5.04 (d, 7.5) |
| 3′′ | 49.8 | 4.36 (m) | |||
| 4′′ | 16.5 | 1.10 (m) | |||
| 5′′ | 167.8 | - | |||
| 1′′′ | 172.3 | - | |||
| 2′′′ | 52.6 | 4.45 (m) | |||
| 3′′′ | 34.2 | 3.62 (m) 3.26 (m) | |||
| 4′′′ | 170.3 | - | |||
| 5′′′ | 23.2 | 1.85 (s) | |||
| NH2 | - | 8.30 | |||
| Position | 𝛅(13C) | 𝛅(1H), (Mult, J in Hz) | Position | 𝛅(13C) | 𝛅(1H), (Mult, J in Hz) |
|---|---|---|---|---|---|
| 1 | n.d. | - | 1′ | 98.5 | 4.86 (br d, 3.7) |
| 2 | 99.9 | - | 2′ | 30.6 | 2.04 (m) 1.78 (m) |
| 3 | 159.9 | - | 3′ | 75.0 | 3.44 (m) |
| 4 | 189.2 | - | 3′α | 57.1 | 3.24 (s) |
| 5 | 48.7 | 3.26 (m) 3.00 (d, 16.2) | 4′ | 73.9 | - |
| 6 | 78.2 | - | 5′ | 67.3 | 4.41 (m) |
| 7 | 198.7 | - | 6′ | 16.3 | 1.10 (m) |
| 8 | 77.6 | 3.62 (d, 9.8) | 7′ | 69.8 | 5.27 (quart., 6.8) |
| 9 | 69.1 | 3.45 (m) | 8′ | 16.4 | 1.18 (d, 6.7) |
| 10 | 74.5 | 4.02(br t) | 9′ | 176.2 | - |
| 11 | 70.4 | 4.58 (dd, 10.1, 2.0) | 10′ | 34.3 | 2.55 (m) |
| 12 | 72.0 | 3.97 (m) | 11′ | 19.5 | 1.08 (m) |
| 13 | 62.8 | 3.75 (m) | 12′ | 19.6 | 1.12 (m) |
| 14 | 170.9 | - | 1′′ | 169.3 | - |
| 15 | 21.1 | 1.97 (s) | 2′′ | 83.1 | 4.90 (d, 4.7) |
| 3′′ | 51.0 | 4.25 (m) | |||
| 4′′ | 22.5 | 1.38 (m) | |||
| 5′′ | 167.9 | - | |||
| 1′′′ | 172.2 | - | |||
| 2′′′ | 52.2 | 4.51 (m) | |||
| 3′′′ | 34.3 | 3.62 (m) 3.23 (m) | |||
| 4′′′ | 170.2 | - | |||
| 5′′′ | 23.1 | 1.85 (s) | |||
| Position | 𝛅(13C) | 𝛅(1H), (Mult, J in Hz) | Position | 𝛅(13C) | 𝛅(1H), (Mult, J in Hz) |
|---|---|---|---|---|---|
| 1 | n.d. | - | 1′ | 98.4 | 4.89 (br d, 3.5) |
| 2 | n.d. | - | 2′ | 30.7 | 2.04 (m) 1.79 (td, 12.3, 3.6) |
| 3 | 159.9 | - | 3′ | 75.0 | 3.43 (m) |
| 4 | 189.2 | - | 3′α | 57.1 | 3.24 (s) |
| 5 | 48.9 | 3.29 (m) 2.98 (d, 16.2) | 4′ | 73.9 | - |
| 6 | 78.2 | - | 5′ | 67.5 | 4.41 (m) |
| 7 | 198.7 | - | 6′ | 16.5 | 1.10 (m) |
| 8 | 77.7 | 3.64 (d, 10.1) | 7′ | 69.8 | 5.27 (quart., 6.8) |
| 9 | 69.2 | 3.48 (m) | 8′ | 16.4 | 1.18 (d, 6.7) |
| 10 | 74.5 | 4.08 (br t) | 9′ | 175.9 | - |
| 11 | 70.7 | 4.66 (dd, 10.2, 1.8) | 10′ | 41.5 | 2.38 (m) |
| 12 | 71.9 | 4.00 (dt, 9.9, 3.4) | 11′ | 26.9 | 1.60 (m) 1.45 (m) |
| 13 | 62.9 | 3.77 (m) | 12′ | 12.2 | 0.88 (t, 7.0) |
| 14 | 170.8 | - | 13′ | 17.5 | 1.09 |
| 15 | 21.2 | 1.96 (s) | 1′′ | 168.5 | - |
| 2′′ | 80.2 | 5.04 (d, 7.5) | |||
| 3′′ | 49.8 | 4.36 (m) | |||
| 4′′ | 16.5 | 1.10 (m) | |||
| 5′′ | 167.8 | - | |||
| 1′′′ | 172.2 | - | |||
| 2′′′ | 52.6 | 4.46 (m) | |||
| 3′′′ | 34.2 | 3.62 (m) 3.26 (m) | |||
| 4′′′ | 170.3 | - | |||
| 5′′′ | 23.2 | 1.85 (s) | |||
| Position | 𝛅(13C) | 𝛅(1H), (Mult, J in Hz) | Position | 𝛅(13C) | 𝛅(1H), (Mult, J in Hz) |
|---|---|---|---|---|---|
| 1 | n.d. | - | 1′ | 98.4 | 4.89 (m) |
| 2 | 100.0 | - | 2′ | 30.7 | 2.05 (m) 1.79 (m) |
| 3 | 160.0 | - | 3′ | 75.0 | 3.44 (m) |
| 4 | 189.2 | - | 3′α | 57.1 | 3.24 (s) |
| 5 | 48.9 | 3.27 (m) 2.99 (m) | 4′ | 73.9 | - |
| 6 | 78.3 | - | 5′ | 67.4 | 4.41 (m) |
| 7 | 198.7 | - | 6′ | 16.4 | 1.10 (m) |
| 8 | 77.6 | 3.62 (d, 9.8) | 7′ | 69.8 | 5.27 (quart., 6.8) |
| 9 | 69.2 | 3.46 (m) | 8′ | 16.4 | 1.18 (d, 6.7) |
| 10 | 74.6 | 4.01(br t) | 9′ | 175.9 | - |
| 11 | 70.5 | 4.58 (dd, 10.1, 2.0) | 10′ | 41.5 | 2.37 (m) |
| 12 | 72.0 | 3.97 (m) | 11′ | 26.9 | 1.60 (m) 1.44 (m) |
| 13 | 63.0 | 3.76 (m) | 12′ | 12.2 | 0.88 (t, 7.0) |
| 14 | 171.0 | - | 13′ | 17.6 | 1.10 (d, 6.8) |
| 15 | 21.2 | 1.97 (s) | 1′′ | 169.4 | - |
| 2′′ | 83.2 | 4.90 (d, 4.7) | |||
| 3′′ | 51.1 | 4.25 (m) | |||
| 4′′ | 22.5 | 1.37 (m) | |||
| 5′′ | 168.0 | - | |||
| 1′′′ | 172.3 | - | |||
| 2′′′ | 52.2 | 4.51 (m) | |||
| 3′′′ | 34.3 | 3.62 (m) 3.29 (m) | |||
| 4′′′ | 170.3 | - | |||
| 5′′′ | 23.1 | 1.85 (s) | |||
| MIC (µg/mL) | ||||||
|---|---|---|---|---|---|---|
| Microorganism | 1 | 2 | 3 | 4 | 5 | 6 |
| S. aureus | 75 | 75 | 37.5 | 25 | <2.34 | <2.34 |
| S. epidermidis | 50 | 50 | 18.75 | 12.5 | <2.34 | <2.34 |
| E. coli | 150 | 150 | 75 | 100 | >200 | >200 |
| K. pneumoniae | 150 | 150 | 100 | 100 | >200 | >200 |
| C. albicans | 200 | >200 | 200 | >200 | >200 | >200 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoz, J.F.-D.l.; Méndez, C.; Salas, J.A.; Olano, C. Novel Bioactive Paulomycin Derivatives Produced by Streptomyces albus J1074. Molecules 2017, 22, 1758. https://doi.org/10.3390/molecules22101758
Hoz JF-Dl, Méndez C, Salas JA, Olano C. Novel Bioactive Paulomycin Derivatives Produced by Streptomyces albus J1074. Molecules. 2017; 22(10):1758. https://doi.org/10.3390/molecules22101758
Chicago/Turabian StyleHoz, Jorge Fernández-De la, Carmen Méndez, José A. Salas, and Carlos Olano. 2017. "Novel Bioactive Paulomycin Derivatives Produced by Streptomyces albus J1074" Molecules 22, no. 10: 1758. https://doi.org/10.3390/molecules22101758
APA StyleHoz, J. F.-D. l., Méndez, C., Salas, J. A., & Olano, C. (2017). Novel Bioactive Paulomycin Derivatives Produced by Streptomyces albus J1074. Molecules, 22(10), 1758. https://doi.org/10.3390/molecules22101758