5-Aza-2′-Deoxycytidine and Valproic Acid in Combination with CHIR99021 and A83-01 Induce Pluripotency Genes Expression in Human Adult Somatic Cells
Abstract
:1. Introduction
2. Results
2.1. Viability Curves in Fibroblasts by Epigenetic Molecules
2.2. Morphological Changes by Epigenetic Molecules
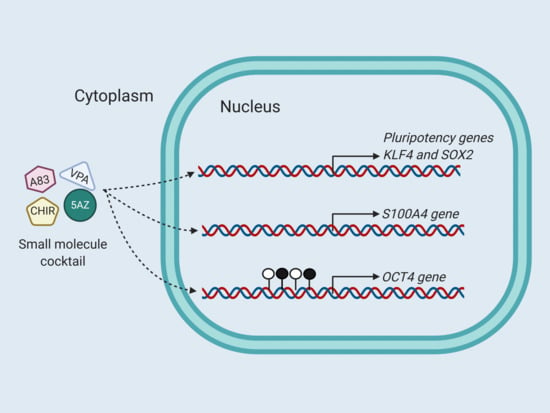
2.3. Expression Analysis of Pluripotent Genes by Effect of Epigenetic Molecules
2.4. Methylation Analysis of OCT4 Promoter by Effect of Epigenetic Molecules
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Reprogramming of BJ Foreskin Neonatal Human Fibroblasts
4.4. HDF Treatment With VPA, 5AZ, and Small Molecules
4.5. Viability Assays
4.6. RNA Extraction
4.7. Reverse Transcription and quantitative PCR Assays
4.8. Bisulfite Sequencing
4.9. Immunofluorescence Assays
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, K.; Yamanaka, S. Induced pluripotent stem cells in medicine and biology. Development 2013, 140, 2457–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Medvedev, S.P.; Shevchenko, A.I.; Zakian, S.M. Induced Pluripotent Stem Cells: Problems and Advantages when Applying them in Regenerative Medicine. Acta Nat. 2010, 2, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Galat, V.; Galat, Y.; Perepitchka, M.; Jennings, L.J.; Iannaccone, P.M.; Hendrix, M.J. Transgene Reactivation in Induced Pluripotent Stem Cell Derivatives and Reversion to Pluripotency of Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells. Stem Cells Dev. 2016, 25, 1060–1072. [Google Scholar] [CrossRef] [Green Version]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; DeVine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K.; et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef]
- Mattout, A.; Biran, A.; Meshorer, E. Global epigenetic changes during somatic cell reprogramming to iPS cells. J. Mol. Cell Biol. 2011, 3, 341–350. [Google Scholar] [CrossRef]
- Weinhold, B. Epigenetics: The science of change. Environ. Health Perspect. 2006, 114, A160–A167. [Google Scholar] [CrossRef] [Green Version]
- Csoka, A.B.; Szyf, M. Epigenetic side-effects of common pharmaceuticals: A potential new field in medicine and pharmacology. Med. Hypotheses 2009, 73, 770–780. [Google Scholar] [CrossRef]
- Christman, J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483–5495. [Google Scholar] [CrossRef] [Green Version]
- Seelan, R.S.; Mukhopadhyay, P.; Pisano, M.M.; Greene, R.M. Effects of 5-Aza-2′-deoxycytidine (decitabine) on gene expression. Drug Metab. Rev. 2018, 50, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Marchion, D.C.; Bicaku, E.; Daud, A.I.; Sullivan, D.M.; Munster, P.N. Valproic acid alters chromatin structure by regulation of chromatin modulation proteins. Cancer Res. 2005, 65, 3815–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phiel, C.J.; Zhang, F.; Huang, E.Y.; Guenther, M.G.; Lazar, M.A.; Klein, P.S. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 2001, 276, 36734–36741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Zhai, Y.; Yu, D.; Cui, J.; Hu, J.F.; Li, W. Valproic Acid Enhances iPSC Induction From Human Bone Marrow-Derived Cells Through the Suppression of Reprogramming-Induced Senescence. J. Cell Physiol. 2016, 231, 1719–1727. [Google Scholar] [CrossRef]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, P.H.; Brick, D.J.; Nethercott, H.E.; Stover, A.E. Traditional human embryonic stem cell culture. Methods Mol. Biol. 2011, 767, 107–123. [Google Scholar]
- Biswas, D.; Jiang, P. Chemically Induced Reprogramming of Somatic Cells to Pluripotent Stem Cells and Neural Cells. Int. J. Mol. Sci. 2016, 17, 226. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhou, H.; Abujarour, R.; Zhu, S.; Young Joo, J.; Lin, T.; Hao, E.; Scholer, H.R.; Hayek, A.; Ding, S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells 2009, 27, 2992–3000. [Google Scholar] [PubMed] [Green Version]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 2010, 7, 651–655. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhao, T.; Guan, J.; Zhang, X.; Fu, Y.; Ye, J.; Zhu, J.; Meng, G.; Ge, J.; Yang, S.; et al. A XEN-like State Bridges Somatic Cells to Pluripotency during Chemical Reprogramming. Cell 2015, 163, 1678–1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dadheech, N.; Srivastava, A.; Belani, M.; Gupta, S.; Pal, R.; Bhonde, R.R.; Srivastava, A.S.; Gupta, S. Basal expression of pluripotency-associated genes can contribute to stemness property and differentiation potential. Stem Cells Dev. 2013, 22, 1802–1817. [Google Scholar] [CrossRef] [Green Version]
- Page, R.L.; Ambady, S.; Holmes, W.F.; Vilner, L.; Kole, D.; Kashpur, O.; Huntress, V.; Vojtic, I.; Whitton, H.; Dominko, T. Induction of stem cell gene expression in adult human fibroblasts without transgenes. Cloning Stem Cells 2009, 11, 417–426. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huangfu, D.; Osafune, K.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Chen, S.; Muhlestein, W.; Melton, D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008, 26, 1269–1275. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Wang, Y.; Zhang, D.; Wang, Y.; Guo, Z.; Zhang, Y. Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology 2008, 70, 622–630. [Google Scholar] [CrossRef]
- Hattori, N.; Nishino, K.; Ko, Y.G.; Hattori, N.; Ohgane, J.; Tanaka, S.; Shiota, K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004, 279, 17063–17069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iglesias, J.M.; Gumuzio, J.; Martin, A.G. Linking Pluripotency Reprogramming and Cancer. Stem Cells Transl. Med. 2017, 6, 335–339. [Google Scholar] [CrossRef]
- Shi, J.; Shi, W.; Ni, L.; Xu, X.; Su, X.; Xia, L.; Xu, F.; Chen, J.; Zhu, J. OCT4 is epigenetically regulated by DNA hypomethylation of promoter and exon in primary gliomas. Oncol. Rep. 2013, 30, 201–206. [Google Scholar] [CrossRef]
- Cao, L.; Li, C.; Shen, S.; Yan, Y.; Ji, W.; Wang, J.; Qian, H.; Jiang, X.; Li, Z.; Wu, M.; et al. OCT4 increases BIRC5 and CCND1 expression and promotes cancer progression in hepatocellular carcinoma. BMC Cancer 2013, 13, 82. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.Y.; Wang, L.T.; Hsu, S.H.; Wang, S.N. Homeobox Genes and Hepatocellular Carcinoma. Cancers (Basel) 2019, 11, 621. [Google Scholar] [CrossRef] [Green Version]
- Rodda, D.J.; Chew, J.L.; Lim, L.H.; Loh, Y.H.; Wang, B.; Ng, H.H.; Robson, P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005, 280, 24731–24737. [Google Scholar] [CrossRef] [Green Version]
- Marson, A.; Foreman, R.; Chevalier, B.; Bilodeau, S.; Kahn, M.; Young, R.A.; Jaenisch, R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell 2008, 3, 132–135. [Google Scholar] [CrossRef] [Green Version]
- Ai, Z.; Shao, J.; Wu, Y.; Yu, M.; Du, J.; Shi, X.; Shi, X.; Zhang, Y.; Guo, Z. CHIR99021 enhances Klf4 Expression through beta-Catenin Signaling and miR-7a Regulation in J1 Mouse Embryonic Stem Cells. PLoS ONE 2016, 11, e0150936. [Google Scholar] [CrossRef] [Green Version]
- Dozmorov, I.; Knowlton, N.; Tang, Y.; Shields, A.; Pathipvanich, P.; Jarvis, J.N.; Centola, M. Hypervariable genes--experimental error or hidden dynamics. Nucleic Acids Res. 2004, 32, e147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, R.J.; Liu, Y.; Marles, L.; Yang, Z.; Trempus, C.; Li, S.; Lin, J.S.; Sawicki, J.A.; Cotsarelis, G. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 2004, 22, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Bian, Y.; Wang, Y.; Chen, L.; Yu, A.; Sun, X. S100A4 influences cancer stem cell-like properties of MGC803 gastric cancer cells by regulating GDF15 expression. Int. J. Oncol. 2016, 49, 559–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Zhou, Y.; Zhou, X.; Guo, Y.; Huang, D.; Zhang, J.; Wang, C.; Cai, L. S100A4 suppresses cancer stem cell proliferation via interaction with the IKK/NF-kappaB signaling pathway. BMC Cancer 2018, 18, 763. [Google Scholar] [CrossRef]
- Chow, K.H.; Park, H.J.; George, J.; Yamamoto, K.; Gallup, A.D.; Graber, J.H.; Chen, Y.; Jiang, W.; Steindler, D.A.; Neilson, E.G.; et al. S100A4 Is a Biomarker and Regulator of Glioma Stem Cells That Is Critical for Mesenchymal Transition in Glioblastoma. Cancer Res. 2017, 77, 5360–5373. [Google Scholar] [CrossRef] [Green Version]
- Flavahan, W.A.; Wu, Q.; Hitomi, M.; Rahim, N.; Kim, Y.; Sloan, A.E.; Weil, R.J.; Nakano, I.; Sarkaria, J.N.; Stringer, B.W.; et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat. Neurosci. 2013, 16, 1373–1382. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef]
- Shi, Y.; Ou, L.; Han, S.; Li, M.; Pena, M.M.; Pena, E.A.; Liu, C.; Nagarkatti, M.; Fan, D.; Ai, W. Deficiency of Kruppel-like factor KLF4 in myeloid-derived suppressor cells inhibits tumor pulmonary metastasis in mice accompanied by decreased fibrocytes. Oncogenesis 2014, 3, e129. [Google Scholar] [CrossRef] [Green Version]
- Stein, U.; Arlt, F.; Walther, W.; Smith, J.; Waldman, T.; Harris, E.D.; Mertins, S.D.; Heizmann, C.W.; Allard, D.; Birchmeier, W.; et al. The metastasis-associated gene S100A4 is a novel target of beta-catenin/T-cell factor signaling in colon cancer. Gastroenterology 2006, 131, 1486–1500. [Google Scholar] [CrossRef]
- Schneider, M.; Hansen, J.L.; Sheikh, S.P. S100A4: A common mediator of epithelial-mesenchymal transition, fibrosis and regeneration in diseases? J. Mol. Med. 2008, 86, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, M.; Zhou, Y.; Wang, F.; Li, X.; Wang, L.; Fan, Q. S100A4 participates in epithelial-mesenchymal transition in breast cancer via targeting MMP2. Tumour. Biol. 2016, 37, 2925–2932. [Google Scholar] [CrossRef] [PubMed]
- Ning, Q.; Li, F.; Wang, L.; Li, H.; Yao, Y.; Hu, T.; Sun, Z. S100A4 amplifies TGF-beta-induced epithelial-mesenchymal transition in a pleural mesothelial cell line. J. Investig. Med. 2018, 66, 334–339. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Shi, J.; Xu, Z.; Yao, X.; Mou, T.; Yu, J.; Liu, H.; Li, G. S100A4-MYH9 Axis Promote Migration and Invasion of Gastric Cancer Cells by Inducing TGF-beta-Mediated Epithelial-Mesenchymal Transition. J. Cancer 2018, 9, 3839–3849. [Google Scholar] [CrossRef] [Green Version]
- Hua, T.; Liu, S.; Xin, X.; Cai, L.; Shi, R.; Chi, S.; Feng, D.; Wang, H. S100A4 promotes endometrial cancer progress through epithelial-mesenchymal transition regulation. Oncol. Rep. 2016, 35, 3419–3426. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguirre-Vázquez, A.; Salazar-Olivo, L.A.; Flores-Ponce, X.; Arriaga-Guerrero, A.L.; Garza-Rodríguez, D.; Camacho-Moll, M.E.; Velasco, I.; Castorena-Torres, F.; Dadheech, N.; Bermúdez de León, M. 5-Aza-2′-Deoxycytidine and Valproic Acid in Combination with CHIR99021 and A83-01 Induce Pluripotency Genes Expression in Human Adult Somatic Cells. Molecules 2021, 26, 1909. https://doi.org/10.3390/molecules26071909
Aguirre-Vázquez A, Salazar-Olivo LA, Flores-Ponce X, Arriaga-Guerrero AL, Garza-Rodríguez D, Camacho-Moll ME, Velasco I, Castorena-Torres F, Dadheech N, Bermúdez de León M. 5-Aza-2′-Deoxycytidine and Valproic Acid in Combination with CHIR99021 and A83-01 Induce Pluripotency Genes Expression in Human Adult Somatic Cells. Molecules. 2021; 26(7):1909. https://doi.org/10.3390/molecules26071909
Chicago/Turabian StyleAguirre-Vázquez, Alain, Luis A. Salazar-Olivo, Xóchitl Flores-Ponce, Ana L. Arriaga-Guerrero, Dariela Garza-Rodríguez, María E. Camacho-Moll, Iván Velasco, Fabiola Castorena-Torres, Nidheesh Dadheech, and Mario Bermúdez de León. 2021. "5-Aza-2′-Deoxycytidine and Valproic Acid in Combination with CHIR99021 and A83-01 Induce Pluripotency Genes Expression in Human Adult Somatic Cells" Molecules 26, no. 7: 1909. https://doi.org/10.3390/molecules26071909
APA StyleAguirre-Vázquez, A., Salazar-Olivo, L. A., Flores-Ponce, X., Arriaga-Guerrero, A. L., Garza-Rodríguez, D., Camacho-Moll, M. E., Velasco, I., Castorena-Torres, F., Dadheech, N., & Bermúdez de León, M. (2021). 5-Aza-2′-Deoxycytidine and Valproic Acid in Combination with CHIR99021 and A83-01 Induce Pluripotency Genes Expression in Human Adult Somatic Cells. Molecules, 26(7), 1909. https://doi.org/10.3390/molecules26071909