Functional and Biochemical Characterization of Three Recombinant Human Glucose-6-Phosphate Dehydrogenase Mutants: Zacatecas, Vanua-Lava and Viangchan
Abstract
:1. Introduction
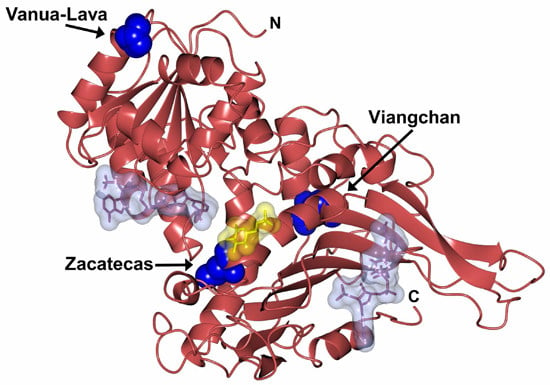
2. Results and Discussion
2.1. Plasmid Construction and Heterologous Expression of Glucose-6-Phosphate Dehydrogenase (G6PD)
2.2. Purification and Characterization of Recombinant G6PDs
2.3. Functional Characterization
2.4. Evaluation of Protein Stability
2.5. Spectroscopic Characterization
3. Materials and Methods
3.1. Gene Synthesis and Plasmid Construction
3.2. Expression and Purification
3.3. Functional Characterization
3.4. Evaluation of Protein Stability
3.5. Spectroscopic Characterization
3.6. In Silico Mutagenesis and Modeling
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pai, G.S.; Sprenkle, J.A.; Do, T.T.; Mareni, C.E.; Migeon, B.R. Localization of loci for hypoxanthine phosphoribosyltransferase and glucose-6-phosphate dehydrogenase and biochemical evidence of nonrandom X chromosome expression from studies of a human X-autosome translocation. Proc. Natl. Acad. Sci. USA 1980, 77, 2810–2813. [Google Scholar] [CrossRef] [PubMed]
- Mason, P.J.; Bautista, J.M.; Gilsanz, F. G6PD deficiency: The genotype-phenotype association. Blood Rev. 2007, 5, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Nkhoma, E.T.; Poole, C.; Vannappagari, V.; Hall, S.A.; Beutler, E. The global prevalence of glucose-6-phosphate dehydrogenase deficiency: A systematic review and meta-analysis. Blood Cells Mol. Dis. 2009, 42, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Beutler, E. G6PD deficiency. Blood 1994, 84, 3613–3636. [Google Scholar] [PubMed]
- Cappellini, D.; Fiorelli, G. Glucose-6-phosphate dehydrogenase deficiency. Lancet 2008, 606, 64–74. [Google Scholar] [CrossRef]
- Minucci, B.; Giardina, C.; Zuppi, E. Glucose-6-phosphate dehydrogenase laboratory assay: How, when, and why? IUBMB Life 2009, 61, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Minucci, A.; Moradkhani, K.; Hwang, M.; Zuppi, C.; Giardina, B.; Capoluongo, P. Glucose-6-phosphate dehydrogenase (G6PD) mutations database: Review of the “old” and update of the new mutations. Blood Cells Mol. Dis. 2012, 48, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.J.; Martin, A.C.; Au, S.W.; Lam, V.M. G6PDdb, an integrated database of glucose-6-phosphate dehydrogenase (G6PD) mutations. Hum. Mutat. 2002, 19, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Vaca, G.; Arambula, E.; Esparza, A. Molecular heterogeneity of glucose-6-phosphate dehydrogenase deficiency in Mexico: Overall results of a 7-year project. Blood Cells Mol. Dis. 2002, 28, 436–444. [Google Scholar] [CrossRef] [PubMed]
- García-Magallanes, N.; Luque-Ortega, F.; Aguilar-Medina, E.M.; Ramos-Payán, R.; Galaviz-Hernández, C.; Romero-Quintana, J.G.; Del Pozo-Yauner, L.; Rangel-Villalobos, H.; Arámbula-Meraz, E.J. Glucose-6-phosphate dehydrogenase deficiency in northern Mexico and description of a novel mutation. J. Genet. 2014, 93, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Vaca, G.; Arámbula, E.; Monsalvo, A.; Medina, C.; Nuñez, C.; Sandoval, L.; López-Guido, B. Glucose-6-phosphate dehydrogenase (G6PD) mutations in Mexico: Four new G6PD variants. Blood Cells Mol. Dis. 2003, 31, 112–120. [Google Scholar] [CrossRef]
- Chui, D.H.D.; Wong, W.C.; Chung, S.W.; Patterson, M.; Bhargava, S.; Pooh, M.C. Embryonic α-globin chains in adults: A marker for athalassemia-1 haplotype due to a 17.5-kb deletion. N. Engl. J. Med. 1986, 314, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Beutler, E.; Kuhl, W.; German, F.; Sdenz, R.; Rodriguez, W.R. Mutation analysis of glucose-6-phosphate dehydrogenase (G6PD) variants in Costa Rica. Hum. Genet. 1991, 87, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Westwood, B.; Bartsocas, C.S.; Malcorra-Azpiazu, J.J.; Indrak, K.; Beutler, E. Glucose-6-phosphate dehydrogenase mutations and haplotypes in various ethnic groups. Blood 1995, 85, 257–263. [Google Scholar] [PubMed]
- Iwai, K.; Hirono, A.; Matsuoka, H.; Kawamoto, F.; Horie, T.; Lin, K.; Tanlular, I.S.; Dachlan, Y.P.; Notopuro, H.; Hidayah, N.I.; et al. Distribution of glucose-6-phosphate dehydrogenase mutations in Southeast Asia. Hum. Genet. 2001, 108, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Nuchprayoon, I.; Sanpavat, S.; Nuchprayoon, S. Glucose-6-phosphate dehydrogenase (G6PD) mutations in Thailand: G6PD Viangchan (871 G > A) is the most common deficient variant in the Thai population. Hum. Mutat. 2002, 19, 185. [Google Scholar] [CrossRef] [PubMed]
- Kumakawa, T.; Suzuki, S.; Fujii, H.; Miwa, S. Frequency of glucose 6-phosphate dehydrogenase (G6PD) deficiency in Tokyo and a new variant: G6PD Musashino. Nippon Ketsueki Gakkai Zasshi 1987, 50, 25–28. [Google Scholar] [PubMed]
- McNicholas, S.; Potterton, E.; Wilson, K.S.; Noble, M.E.M. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Manzo, S.; Terrón-Hernández, J.; de la Mora-de la Mora, I.; García Torres, I.; López-Velázquez, G.; Reyes-Vivas, H.; Oria-Hernández, J. Cloning, expression, purification and characterization of His-tagged human glucose-6-phosphate dehydrogenase: A simplified method for protein yield. Protein J. 2013, 32, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Lam, V.M.; Engel, P.C. Marked decrease in specific activity contributes to disease phenotype in two human glucose-6-phosphate dehydrogenase mutants, G6PDUnion and G6PDAndalus. Hum. Mutat. 2005, 26, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Lam, V.M.S.; Engel, P.C. Functional properties of two mutants of human glucose 6-phosphate dehydrogenase, R393G and R393H, corresponding to the clinical variants G6PD Wisconsin and Nashville. Biochim. Biophys. Acta 2006, 1762, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Choi, M.Y.; Au, S.W.; Au, D.M.; Lam, V.M.S.; Engel, P.C. Purification and detailed study of two clinically different human glucose 6-phosphate dehydrogenase variants, G6PD (Plymouth) and G6PD (Mahidol): Evidence for defective protein folding as the basis of disease. Mol. Genet. Metab. 2008, 93, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Manzo, S.; Terrón-Hernández, J.; De la Mora-De la Mora, I.; González-Valdez, A.; Marcial-Quino, J.; García-Torres, I.; Vanoye-Carlo, A.; López-Velázquez, G.; Hernández-Alcantara, G.; Oria-Hernández, J.; et al. The stability of G6PD is affected by mutations with different clinical phenotypes. Int. J. Mol. Sci. 2014, 15, 21179–21201. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Manzo, S.; Marcial-Quino, J.; Vanoye-Carlo, A.; Enríquez-Flores, S.; De la Mora-De la Mora, I.; González-Valdez, A.; García-Torres, I.; Martínez-Rosas, V.; Sierra-Palacios, E.; Lazcano-Pérez, F.; et al. Mutations of glucose-6-phosphate dehydrogenase durham, Santa-Maria and A+ variants are associated with loss functional and structural stability of the protein. Int. J. Mol. Sci. 2015, 16, 28657–28668. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Chan, T.F.; Lam, V.; Engel, P. What is the role of the second “structural” NADP-binding site in human glucose-6-phosphate dehydrogenase? Protein Sci. 2008, 17, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Kiani, F.; Schwarzl, S.; Fischer, S.; Efferth, T. Three-dimensional modeling of glucose-6-phosphate dehydrogenase-deficient variants from German ancestry. PLoS ONE 2007, 7, e625. [Google Scholar]
- Man-Chiu, P.; Kelley, H.; William, C.S.; Prchal, J.T. G6PD Viangchan: A new glucose 6-phosphate dehydrogenase variant from Laos. Hum. Genet. 1998, 78, 98–99. [Google Scholar]
- Boonyuen, U.; Chamchoy, K.; Swangsri, T.; Saralamba, T.; Day, N.P.J.; Imwong, M. Detailed functional analysis of two clinical glucose-6-phosphate dehydrogenase (G6PD) variants, G6PDViangchan and G6PDViangchan + Mahidol: Decreased stability and catalytic efficiency contribute to the clinical phenotype. Mol. Genet. Metab. 2016. [Google Scholar] [CrossRef] [PubMed]
- Beutler, E. The genetics of glucose-6-phosphate dehydro-genase deficiency. Semin. Hematol. 1990, 27, 137–164. [Google Scholar] [PubMed]
- Scopes, D.A.; Bautista, J.M.; Naylor, C.E.; Adams, M.J.; Mason, P.J. Amino acid substitutions at the dimer interface of human glucose-6-phosphate dehydrogenase that increase thermostability and reduce the stabilising effect of NADP. Eur. J. Biochem. 1998, 251, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Engel, P.C. Clinical mutants of human glucose-6-phosphate dehydrogenase: Impairment of NADP+ binding affects both folding and stability. Biochim. Biophys. Acta 2009, 1792, 804–809. [Google Scholar] [CrossRef] [PubMed]
- Au, S.W.N.; Gover, S.; Lam, V.M.S.; Adams, M. Human glucose-6-phosphate dehydrogenase: The crystal structure reveals a structural NADP+ molecule and provides insights into enzyme deficiency. Structure 2000, 8, 293–303. [Google Scholar] [CrossRef]
- Verma, A.; Chandra, S.; Suthar, M.K.; Doharey, P.K.; Siddiqi, M.I.; Saxena, J.K. NADP+ binding effects tryptophan accessibility, folding and stability of recombinant B. malayi G6PD. Int. J. Biol. Macromol. 2016, 85, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct. Funct. Bioinform. 2009, 77, 114–122. [Google Scholar] [CrossRef] [PubMed]










| G6PD Enzyme | Total Protein (mg) | Specific Activity (IU·mg−1) | Total Activity (IU) | Yield (%) |
|---|---|---|---|---|
| Wild Type | 2.1 | 230 | 483 | 61 |
| Zacatecas | 1.46 | 58 | 85 | 40 |
| Vanua-Lava | 0.5 | 182 | 91 | 42 |
| Viangchan | 1.32 | 228 | 300 | 43 |
| Kinetic Constants | WT-G6PD | Mutants | ||
|---|---|---|---|---|
| Zacatecas | Vanua-Lava | Viangchan | ||
| KmG6P (µM) | 38.49 | 111 | 34 | 42 |
| KmNADP+ (µM) | 6.16 | 24 | 18 | 17 |
| kcat (s−1) | 230 | 58 | 142 | 145 |
| kcat/Km·G6P (s−1·M−1) | 5.97 × 106 | 0.52 × 106 | 4.17 × 106 | 3.45 × 106 |
| kcat/KmNADP+ (s−1·M−1) | 37.33 × 106 | 2.41 × 106 | 7.88 × 106 | 8.52 × 106 |
| Strain E. coli | Relevant Characteristic(s) or Sequence | Source and/or Reference |
|---|---|---|
| BW25113 | F−, DE(araD-araB)567, lacZ4787(del)::rrnB-3, LAM−, rph-1, DE(rhaD-rhaB)568, hsdR514 | [34] |
| BL21(DE3)Δzwf::kanr | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 (lacI lacUV5-T7 gene 1 ind1 sam7 nin5)) Δzwf-777::kan | [23] |
| Plasmids | ||
| pETg6pd | pET-3a carrying the human g6pd gene, AmpR | [23] |
| pETgR257L | pET-3a carrying the human g6pd gene with a R257L mutation in the G6PD protein, AmpR | This study |
| pETgL128P | pET-3a carrying the human g6pd gene with an L128P mutation in the G6PD protein, AmpR | This study |
| pETgV291M | pET-3a carrying the human g6pd gene with an V291M mutation in the G6PD protein, AmpR | This study |
| Mutagenesis | Primer Sequence | |
| R257L fw | 5′-GGATCATCCTGGACGTGATG-3′ | This study |
| R257L rev | 5′-CATCACGTCCAGGATGATTCC-3′ | This study |
| L128P fw | 5′-GAATGCCCTCCACCTGGG-3′ | This study |
| L128P rev | 5′-CTTACGGGAGGTGGACCC-3′ | This study |
| V291M fw | 5′-GAGAAGGTCAAGATGTTGAAATG-3′ | This study |
| V291M rev | 5′-CATTTCAACATCTTCACCTTCTC-3′ | This study |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Manzo, S.; Marcial-Quino, J.; Vanoye-Carlo, A.; Serrano-Posada, H.; González-Valdez, A.; Martínez-Rosas, V.; Hernández-Ochoa, B.; Sierra-Palacios, E.; Castillo-Rodríguez, R.A.; Cuevas-Cruz, M.; et al. Functional and Biochemical Characterization of Three Recombinant Human Glucose-6-Phosphate Dehydrogenase Mutants: Zacatecas, Vanua-Lava and Viangchan. Int. J. Mol. Sci. 2016, 17, 787. https://doi.org/10.3390/ijms17050787
Gómez-Manzo S, Marcial-Quino J, Vanoye-Carlo A, Serrano-Posada H, González-Valdez A, Martínez-Rosas V, Hernández-Ochoa B, Sierra-Palacios E, Castillo-Rodríguez RA, Cuevas-Cruz M, et al. Functional and Biochemical Characterization of Three Recombinant Human Glucose-6-Phosphate Dehydrogenase Mutants: Zacatecas, Vanua-Lava and Viangchan. International Journal of Molecular Sciences. 2016; 17(5):787. https://doi.org/10.3390/ijms17050787
Chicago/Turabian StyleGómez-Manzo, Saúl, Jaime Marcial-Quino, America Vanoye-Carlo, Hugo Serrano-Posada, Abigail González-Valdez, Víctor Martínez-Rosas, Beatriz Hernández-Ochoa, Edgar Sierra-Palacios, Rosa Angélica Castillo-Rodríguez, Miguel Cuevas-Cruz, and et al. 2016. "Functional and Biochemical Characterization of Three Recombinant Human Glucose-6-Phosphate Dehydrogenase Mutants: Zacatecas, Vanua-Lava and Viangchan" International Journal of Molecular Sciences 17, no. 5: 787. https://doi.org/10.3390/ijms17050787
APA StyleGómez-Manzo, S., Marcial-Quino, J., Vanoye-Carlo, A., Serrano-Posada, H., González-Valdez, A., Martínez-Rosas, V., Hernández-Ochoa, B., Sierra-Palacios, E., Castillo-Rodríguez, R. A., Cuevas-Cruz, M., Rodríguez-Bustamante, E., & Arreguin-Espinosa, R. (2016). Functional and Biochemical Characterization of Three Recombinant Human Glucose-6-Phosphate Dehydrogenase Mutants: Zacatecas, Vanua-Lava and Viangchan. International Journal of Molecular Sciences, 17(5), 787. https://doi.org/10.3390/ijms17050787