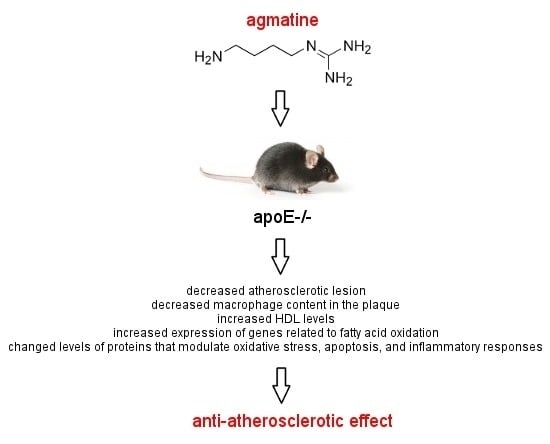
Anti-Atherosclerotic Action of Agmatine in ApoE-Knockout Mice
Abstract
:1. Introduction
2. Results
2.1. Body Weight
2.2. Effects of Agmatine on Atherosclerosis
2.3. RT2 Profiler PCR Arrays
2.4. Influence of Agmatine on Liver Mitoproteome
3. Discussion
3.1. Effect of Agmatine on the Development and Structure of Atherosclerotic Lesions in ApoE-Knockout Mice
3.2. Mechanisms of Anti-Atherosclerotic Action of Agmatine in ApoE-Knockout Mice
3.3. The Effect of Agmatine on mRNA Expression of Factors Involved in Hepatic Lipid Metabolism
3.4. Effect of Agmatine on Liver Mitoproteome
3.5. Conclusion and Future Directions
4. Materials and Methods
4.1. Animal Experiments
4.2. Analysis of Atherosclerotic Plaque
4.3. Biochemical Methods
4.4. RT2 Profiler PCR Arrays
4.5. Two-Dimensional Electrophoresis (2-DE) and Gel Image Analysis
4.6. LC MS/MS
4.7. Immunoblotting
4.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lloyd-Jones, D.M. Cardiovascular risk prediction: Basic concepts, current status, and future directions. Circulation 2010, 121, 1768–1777. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Okamoto, Y.; Rocha, V.Z.; Folco, E. Inflammation in atherosclerosis: Transition from theory to practice. Circ. J. 2010, 74, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Di Lisa, F.; Kaludercic, N.; Carpi, A.; Menabo, R.; Giorgio, M. Mitochondria and vascular pathology. Pharmacol. Rep. 2009, 61, 123–130. [Google Scholar] [CrossRef]
- Gutierrez, J.; Ballinger, S.W.; Darley-Usmar, V.M.; Landar, A. Free radicals, mitochondria, and oxidized lipids: The emerging role in signal transduction in vascular cells. Circ. Res. 2006, 99, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Nassir, F.; Ibdah, J.A. Role of mitochondria in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2014, 15, 8713–8742. [Google Scholar] [CrossRef] [PubMed]
- Karnewar, S.; Vasamsetti, S.B.; Gopoju, R.; Kanugula, A.K.; Ganji, S.K.; Prabhakar, S.; Rangaraj, N.; Tupperwar, N.; Kumar, J.M.; Kotamraju, S. Mitochondria-targeted esculetin alleviates mitochondrial dysfunction by AMPK-mediated nitric oxide and SIRT3 regulation in endothelial cells: Potential implications in atherosclerosis. Sci. Rep. 2016, 6, 24108. [Google Scholar] [CrossRef] [PubMed]
- Agostinelli, E.; Marques, M.P.; Calheiros, R.; Gil, F.P.; Tempera, G.; Viceconte, N.; Battaglia, V.; Grancara, S.; Toninello, A. Polyamines: Fundamental characters in chemistry and biology. Amino Acids 2010, 38, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Moinard, C.; Cynober, L.; de Bandt, J.P. Polyamines: Metabolism and implications in human diseases. Clin. Nutr. 2005, 24, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Satriano, J. Agmatine: At the crossroads of the arginine pathways. Ann. N. Y. Acad. Sci. 2003, 1009, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Berkels, R.; Taubert, D.; Grundemann, D.; Schomig, E. Agmatine signaling: Odds and threads. Cardiovasc. Drug Rev. 2004, 22, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Head, G.A.; Mayorov, D.N. Imidazoline receptors, novel agents and therapeutic potential. Cardiovasc. Hematol. Agents Med. Chem. 2006, 4, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.W.; Zheng, R.Y. Neuroprotective effects of receptor imidazoline 2 and its endogenous ligand agmatine. Neurosci. Bull. 2006, 22, 187–191. [Google Scholar] [PubMed]
- Arndt, M.A.; Battaglia, V.; Parisi, E.; Lortie, M.J.; Isome, M.; Baskerville, C.; Pizzo, D.P.; Ientile, R.; Colombatto, S.; Toninello, A.; et al. The arginine metabolite agmatine protects mitochondrial function and confers resistance to cellular apoptosis. Am. J. Physiol. Cell Physiol. 2009, 296, C1411–C1419. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, V.; Rossi, C.A.; Colombatto, S.; Grillo, M.A.; Toninello, A. Different behavior of agmatine in liver mitochondria: Inducer of oxidative stress or scavenger of reactive oxygen species? Biochim. Biophys. Acta 2007, 1768, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, V.; Grancara, S.; Satriano, J.; Saccoccio, S.; Agostinelli, E.; Toninello, A. Agmatine prevents the Ca2+-dependent induction of permeability transition in rat brain mitochondria. Amino Acids 2010, 38, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, V.; Grancara, S.; Mancon, M.; Cravanzola, C.; Colombatto, S.; Grillo, M.A.; Tempera, G.; Agostinelli, E.; Toninello, A. Agmatine transport in brain mitochondria: A different mechanism from that in liver mitochondria. Amino Acids 2010, 38, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Nissim, I.; Horyn, O.; Daikhin, Y.; Chen, P.; Li, C.; Wehrli, S.L.; Nissim, I.; Yudkoff, M. The molecular and metabolic influence of long term agmatine consumption. J. Biol. Chem. 2014, 289, 9710–9729. [Google Scholar] [CrossRef] [PubMed]
- Nissim, I.; Daikhin, Y.; Nissim, I.; Luhovyy, B.; Horyn, O.; Wehrli, S.L.; Yudkoff, M. Agmatine stimulates hepatic fatty acid oxidation: A possible mechanism for up-regulation of ureagenesis. J. Biol. Chem. 2006, 281, 8486–8496. [Google Scholar] [CrossRef] [PubMed]
- El Awady, M.S.; Suddek, G.M. Agmatine ameliorates atherosclerosis progression and endothelial dysfunction in high cholesterol-fed rabbits. J. Pharm. Pharmacol. 2014, 66, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Kitajima, S.; Watanabe, T.; Xu, J.; Zhang, J.; Liu, E.; Chen, Y.E. Rabbit models for the study of human atherosclerosis: From pathophysiological mechanisms to translational medicine. Pharmacol. Ther. 2015, 146, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Meir, K.S.; Leitersdorf, E. Atherosclerosis in the apolipoprotein-E-deficient mouse: A decade of progress. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, N.; Hansen, J.; Palmfeldt, J. Mitochondrial proteomics—A tool for the study of metabolic disorders. J. Inherit. Metab. Dis. 2012, 35, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Joy, T.R. Novel HDL-based therapeutic agents. Pharmacol. Ther. 2012, 135, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, P.; Ray, K.K.; Cannon, C.P. High-density lipoprotein and coronary heart disease: Current and future therapies. J. Am. Coll. Cardiol. 2010, 55, 1283–1299. [Google Scholar] [CrossRef] [PubMed]
- Nofer, J.R.; Brodde, M.F.; Kehrel, B.E. High-density lipoproteins, platelets and the pathogenesis of atherosclerosis. Clin. Exp. Pharmacol. Physiol. 2010, 37, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Redondo, S.; Martinez-Gonzalez, J.; Urraca, C.; Tejerina, T. Emerging therapeutic strategies to enhance HDL function. Lipids Health Dis. 2011, 10, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miinalainen, I.J.; Schmitz, W.; Huotari, A.; Autio, K.J.; Soininen, R.; van Themaat, E.V.L.; Baes, M.; Herzig, K.H.; Conzelmann, E.; Hiltunen, J.K. Mitochondrial 2,4-dienoyl-CoA reductase deficiency in mice results in severe hypoglycemia with stress intolerance and unimpaired ketogenesis. PLoS Genet. 2009, 5, e1000543. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M.; Gradinger, A.; Longo, N.; Wu, X.; Leclerc, D.; Lerner-Ellis, J.; Lemieux, M.; Belair, C.; Watkins, D.; Rosenblatt, D.S.; et al. Homozygous nonsense mutation in the MCEE gene and siRNA suppression of methylmalonyl-CoA epimerase expression: A novel cause of mild methylmalonic aciduria. Mol. Genet. Metab. 2006, 88, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, N.; Lopes-Virella, M.F. Oxidized LDL induces the expression of ALBP/aP2 mRNA and protein in human THP-1 macrophages. J. Lipid Res. 2000, 41, 2017–2023. [Google Scholar] [PubMed]
- Kazemi, M.R.; McDonald, C.M.; Shigenaga, J.K.; Grunfeld, C.; Feingold, K.R. Adipocyte fatty acid-binding protein expression and lipid accumulation are increased during activation of murine macrophages by toll-like receptor agonists. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Makowski, L.; Boord, J.B.; Maeda, K.; Babaev, V.R.; Uysal, K.T.; Morgan, M.A.; Parker, R.A.; Suttles, J.; Fazio, S.; Hotamisligil, G.S.; et al. Lack of macrophage fatty-acid-binding protein aP2 protects mice deficient in apolipoprotein E against atherosclerosis. Nat. Med. 2001, 7, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Suhre, K.; Romisch-Margl, W.; de Angelis, M.H.; Adamski, J.; Luippold, G.; Augustin, R. Identification of a potential biomarker for FABP4 inhibition: The power of lipidomics in preclinical drug testing. J. Biomol. Screen. 2011, 16, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Kamanna, V.S.; Kashyap, M.L. Mechanism of action of niacin. Am. J. Cardiol. 2008, 101, 20B–26B. [Google Scholar] [CrossRef] [PubMed]
- Kamanna, V.S.; Ganji, S.H.; Kashyap, M.L. Niacin: An old drug rejuvenated. Curr. Atheroscler. Rep. 2009, 11, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Drzewinska, J.; Pulaski, L.; Soszynski, M.; Bartosz, G. Seladin-1/DHCR24: A key protein of cell homeostasis and cholesterol biosynthesis. Postepy Hig. Med. Dosw. 2009, 63, 318–330. [Google Scholar]
- Jiao, Y.; Zan, L.S.; Liu, Y.F.; Wang, H.B. Molecular characterization, polymorphism of the ACOX1 gene and association with ultrasound traits in Bos taurus. Genet. Mol. Res. 2011, 10, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Infante, J.P.; Tschanz, C.L.; Shaw, N.; Michaud, A.L.; Lawrence, P.; Brenna, J.T. Straight-chain acyl-CoA oxidase knockout mouse accumulates extremely long chain fatty acids from α-linolenic acid: Evidence for runaway carousel-type enzyme kinetics in peroxisomal beta-oxidation diseases. Mol. Genet. Metab. 2002, 75, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Fraccascia, P.; Casteels, M.; de Schryver, E.; van Veldhoven, P.P. Role of thiamine pyrophosphate in oligomerisation, functioning and import of peroxisomal 2-hydroxyacyl-CoA lyase. Biochim. Biophys. Acta 2011, 1814, 1226–1233. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Ohkawa, K. Chemotherapeutic agents that induce mitochondrial apoptosis. Curr. Cancer Drug Targets 2004, 4, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Sasagawa, A.; Takeuchi, H.; Shibata, S.; Marushima, H.; Mamori, S.; Ohkawa, K. Conformational change in the active center region of GST P1–1, due to binding of a synthetic conjugate of DXR with GSH, enhanced JNK-mediated apoptosis. Apoptosis 2007, 12, 1269–1280. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.Y.; Liu, J.; Nikolic-Paterson, D.J. The role of stress-activated protein kinase signaling in renal pathophysiology. Braz. J. Med. Biol. Res. 2009, 42, 29–37. [Google Scholar] [CrossRef] [PubMed]
- York, J.L.; Maddox, L.C.; Zimniak, P.; McHugh, T.E.; Grant, D.F. Reduction of MTT by glutathione S-transferase. Biotechniques 1998, 25, 622–628. [Google Scholar] [PubMed]
- Kraus, S.; Bunsen, T.; Schuster, S.; Cichon, M.A.; Tacke, M.; Reinheckel, T.; Sommerhoff, C.P.; Jochum, M.; Nagler, D.K. Cellular senescence induced by cathepsin X downregulation. Eur. J. Cell Biol. 2011, 90, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, A.A.; Srivatsa, U. Heat-shock protein 60 and cardiovascular disease: A paradoxical role. Future Cardiol. 2008, 4, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Reed, J.C.; Homma, S. Heat-shock proteins as regulators of apoptosis. Oncogene 2003, 22, 9041–9047. [Google Scholar] [CrossRef] [PubMed]
- Morera, S.; Lacombe, M.L.; Xu, Y.; LeBras, G.; Janin, J. X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 å resolution. Structure 1995, 3, 1307–1314. [Google Scholar] [CrossRef]
- Mitterberger, M.C.; Kim, G.; Rostek, U.; Levine, R.L.; Zwerschke, W. Carbonic anhydrase III regulates peroxisome proliferator-activated receptor-γ2. Exp. Cell Res. 2012, 318, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M. Role of regucalcin in maintaining cell homeostasis and function. Int. J. Mol. Med. 2005, 15, 371–389. [Google Scholar] [CrossRef] [PubMed]
- Bartelt, A.; John, C.; Schaltenberg, N.; Berbee, J.F.P.; Worthmann, A.; Cherradi, M.L.; Schlein, C.; Piepenburg, J.; Boon, M.R.; Rinninger, F.; et al. Thermogenic adipocytes promote HDL turnover and reverse cholesterol transport. Nat. Commun. 2017, 8, 15010. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.S.; Wu, H.T.; Cheng, K.C.; Lin, K.C.; Chen, C.T.; Cheng, J.T. A Novel Mechanism for Decreasing Plasma Lipid Level from Imidazoline I-1 Receptor Activation in High Fat Diet-fed Mice. Horm. Metab. Res. 2011, 43, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Stachowicz, A.; Olszanecki, R.; Suski, M.; Wisniewska, A.; Toton-Zuranska, J.; Madej, J.; Jawien, J.; Bialas, M.; Okon, K.; Gajda, M.; et al. Mitochondrial aldehyde dehydrogenase activation by Alda-1 inhibits atherosclerosis and attenuates hepatic steatosis in apolipoprotein E-knockout mice. J. Am. Heart Assoc. 2014, 3, e001329. [Google Scholar] [CrossRef] [PubMed]
- Jawien, J.; Gajda, M.; Olszanecki, R.; Korbut, R. BAY x 1005 attenuates atherosclerosis in apoE/LDLR—Double knockout mice. J. Physiol. Pharmacol. 2007, 58, 583–588. [Google Scholar] [PubMed]
- Pawlowska, M.; Gajda, M.; Pyka-Fosciak, G.; Toton-Zuranska, J.; Niepsuj, A.; Kus, K.; Bujak-Gizycka, B.; Suski, M.; Olszanecki, R.; Jawien, J.; et al. The effect of doxycycline on atherogenesis in apoE-knockout mice. J. Physiol. Pharmacol. 2011, 62, 247–250. [Google Scholar] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Yan, J.X.; Wait, R.; Berkelman, T.; Harry, R.A.; Westbrook, J.A.; Wheeler, C.H.; Dunn, M.J. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 2000, 21, 3666–3672. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]





| TC (mmol/L) | TG (mmol/L) | HDL (mmol/L) | LDL (mmol/L) | |
|---|---|---|---|---|
| control | 13.8 ± 0.3 | 1.4 ± 0.2 | 3.7 ± 0.9 | 8.2 ± 0.8 |
| agmatine | 19 ± 4.1 (p = 0.052) | 1.7 ± 0.6 (NS) | 8.6 ± 1.8 * | 11.7 ± 2.4 (NS) |
| VCAM-1 (ng/mL) | IL-12 (pg/mL) | MCP-1 (pg/mL) | SAA (ng/mL) | |
|---|---|---|---|---|
| control | 9.09 ± 0.34 | 8.95 ± 0.52 | 45.16 ± 6.96 | 84,222 ± 20,109 |
| agmatine | 10.11 ± 1.16 (NS) | 10.57 ± 0.82 * | 64.82 ± 5.65 * | 101,360 ± 54,611 (NS) |
| Gene Name | Description | Fold Change | p Value (t-Test) |
|---|---|---|---|
| Acsbg2 | Acyl-CoA synthetase bubblegum family member 2 | 2.08 | 0.000608 |
| Acsm2 | Acyl-CoA synthetase medium-chain family member 2 | 2.99 | 0.013220 |
| Cpt1a | Carnitine palmitoyltransferase 1a, liver | 2.07 | 0.000061 |
| Decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | 2.47 | 0.041195 |
| Fabp1 | Fatty acid binding protein 1, liver | 3.51 | 0.001004 |
| Fabp2 | Fatty acid binding protein 2, intestinal | 6.53 | 0.019459 |
| Fabp5 | Fatty acid binding protein 5, epidermal | 2.01 | 0.021016 |
| Mcee | Methylmalonyl CoA epimerase | 2.06 | 0.011877 |
| Prkag2 | Protein kinase, AMP-activated, gamma 2 non-catalytic subunit | 2.33 | 0.003263 |
| Acaa1a | Acetyl-Coenzyme A acyltransferase 1A | −2.04 | 0.007525 |
| Fabp4 | Fatty acid binding protein 4, adipocyte | −2.47 | 0.015985 |
| Gene Name | Description | Fold Change | p Value (t-Test) |
|---|---|---|---|
| Abca2 | ATP-binding cassette, sub-family A (ABC1), member 2 | 2.46 | 0 |
| Akr1d1 | Aldo-keto reductase family 1, member D1 | 2.79 | 0.003913 |
| Angptl3 | Angiopoietin-like 3 | 2.37 | 0.001165 |
| Cyb5r3 | Cytochrome b5 reductase 3 | 2.36 | 0.000713 |
| Cyp51 | Cytochrome P450, family 51 | 11.18 | 0.000105 |
| Cyp7a1 | Cytochrome P450, family 7, subfamily a, polypeptide 1 | 3.22 | 0.000004 |
| Dhcr24 | 24-dehydrocholesterol reductase | 2.31 | 0.005083 |
| Dhcr7 | 7-dehydrocholesterol reductase | 4.96 | 0.002081 |
| Ebp | Phenylalkylamine Ca2+ antagonist (emopamil) binding protein | 2.49 | 0.000205 |
| Fdft1 | Farnesyl diphosphate farnesyl transferase 1 | 6.22 | 0.005329 |
| Fdps | Farnesyl diphosphate synthetase | 21.31 | 0.000338 |
| Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | 5.33 | 0.008786 |
| Hmgcs1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | 12.10 | 0.011450 |
| Idi1 | Isopentenyl-diphosphate delta isomerase | 18.94 | 0.002732 |
| Insig1 | Insulin induced gene 1 | 7.87 | 0.011090 |
| Mvd | Mevalonate (diphospho) decarboxylase | 9.78 | 0.008416 |
| Mvk | Mevalonate kinase | 7.08 | 0.000262 |
| Nsdhl | NAD(P) dependent steroid dehydrogenase-like | 11.77 | 0.000001 |
| Osbpl5 | Oxysterol binding protein-like 5 | 2.25 | 0.000091 |
| Pcsk9 | Proprotein convertase subtilisin/kexin type 9 | 10.98 | 0.012371 |
| Pmvk | Phosphomevalonate kinase | 7.94 | 0.001513 |
| Srebf1 | Sterol regulatory element binding transcription factor 1 | 10.17 | 0.004453 |
| Apoa4 | Apolipoprotein A-IV | −6.23 | 0.021159 |
| Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 | −3.28 | 0.000045 |
| Insig2 | Insulin induced gene 2 | −2.78 | 0.000292 |
| Lipe | Lipase, hormone sensitive | −2.05 | 0.004930 |
| No. | Protein | SwissProt Accession Number | Molecular Mass (kDa) | pI | Unique Peptides | Total Peptides | Protein Coverage (%) | Fold Change |
|---|---|---|---|---|---|---|---|---|
| 1 | Regucalcin | RGN_MOUSE | 33.4 | 4.94 | 7 | 10 | 27 | 1.440 |
| 2 | ATP synthase subunit beta, mitochondrial | ATPB_MOUSE | 56.3 | 5.19 | 5 | 8 | 11 | −1.740 |
| 3 | 2-hydroxyacyl-CoA lyase 1 | HACL1_MOUSE | 63.6 | 5.89 | 7 | 12 | 14 | 2.370 |
| 4 | Hydroxymethylglutaryl-CoA synthase, mit | HMCS2_MOUSE | 56.8 | 8.65 | 2 | 4 | 4.1 | −1.320 |
| 5 | Cytochrome b5 | CYB5_MOUSE | 15.2 | 4.96 | 2 | 4 | 16.,4 | 1.680 |
| 6 | Isocitrate dehydrogenase [NADP] | IDHC_MOUSE | 46.6 | 6.48 | 5 | 8 | 13 | 1.970 |
| 7 | Delta-1-pyrroline-5-carboxylate dehydrogenase | AL4A1_MOUSE | 61.8 | 8.58 | 2 | 2 | 2.70 | 2.130 |
| 8 | Urate oxidase | URIC_MOUSE | 35 | 8.48 | 5 | 6 | 17 | 2.650 |
| 9 | Peroxisomal acyl-Coenzyme A oxidase 1 | ACOX1_MOUSE | 74.6 | 8.64 | 5 | 6 | 9.40 | 3.150 |
| 10 | Acyl-Coenzyme A oxidase 1, palmitoyl | A2A850_MOUSE | 74.6 | 8.64 | 6 | 15 | 11 | 2.840 |
| 11 | Urate oxidase | URIC_MOUSE | 35 | 8.48 | 11 | 19 | 33 | 2.180 |
| 12 | 60 kDa heat shock protein, mitochondrial | CH60_MOUSE | 60.9 | 5.91 | 3 | 5 | 5.1 | −1.790 |
| 13 | Voltage-dependent anion-selective channel 1 | VDAC1_MOUSE | 32.3 | 8.55 | 5 | 9 | 20 | 1.510 |
| 14 | ATP synthase subunit alpha, mitochondrial | ATPA_MOUSE | 59.7 | 9.22 | 4 | 8 | 7.10 | 1.980 |
| 15 | Elongation factor 1-alpha 1 | EF1A1_MOUSE | 50.1 | 9.1 | 4 | 4 | 8.00 | 1.650 |
| 16 | Cathepsin Z | CATZ_MOUSE | 34 | 6.13 | 4 | 13 | 17.6 | 2.230 |
| 17 | Ornithine aminotransferase, mitochondrial | OAT_MOUSE | 48.3 | 6,19 | 4 | 8 | 9.8 | −1.860 |
| 18 | Carbamoyl-phosphate synthase [ammonia], mit | CPSM_MOUSE | 164.5 | 6.48 | 4 | 7 | 3.9 | −1.840 |
| 19 | Nucleoside diphosphate kinase B | NDKB_MOUSE | 17.3 | 6.97 | 7 | 13 | 48.,60 | 1.520 |
| 20 | Hydroxyacyl-coenzyme A dehydrogenase | HCDH_MOUSE | 34.4 | 8.76 | 2 | 5 | 6.10 | −1.660 |
| 21 | ES1 protein homolog, mitochondrial | ES1_MOUSE | 28.1 | 9 | 2 | 2 | 6.80 | −1.570 |
| 22 | Carbamoyl-phosphate synthase [ammonia], mit | CPSM_MOUSE | 164.5 | 6.48 | 3 | 4 | 2.40 | −1.610 |
| 23 | Carbamoyl-phosphate synthase [ammonia], mit | CPSM_MOUSE | 164.5 | 6.48 | 4 | 6 | 3.50 | −2.630 |
| 24 | Carbamoyl-phosphate synthase [ammonia], mit | CPSM_MOUSE | 164.5 | 6.48 | 5 | 11 | 5.30 | −1.660 |
| 25 | Carbonic anhydrase 3 | CAH3_MOUSE | 29.3 | 6.97 | 4 | 6 | 15 | 1.200 |
| 26 | Glutamate dehydrogenase 1, mitochondrial | DHE3_MOUSE | 61.3 | 6.71 | 4 | 5 | 7.90 | −1.560 |
| 27 | Glutathione S-transferase P 1 | GSTP1_MOUSE | 23.6 | 8.13 | 3 | 6 | 20 | 1.420 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiśniewska, A.; Olszanecki, R.; Totoń-Żurańska, J.; Kuś, K.; Stachowicz, A.; Suski, M.; Gębska, A.; Gajda, M.; Jawień, J.; Korbut, R. Anti-Atherosclerotic Action of Agmatine in ApoE-Knockout Mice. Int. J. Mol. Sci. 2017, 18, 1706. https://doi.org/10.3390/ijms18081706
Wiśniewska A, Olszanecki R, Totoń-Żurańska J, Kuś K, Stachowicz A, Suski M, Gębska A, Gajda M, Jawień J, Korbut R. Anti-Atherosclerotic Action of Agmatine in ApoE-Knockout Mice. International Journal of Molecular Sciences. 2017; 18(8):1706. https://doi.org/10.3390/ijms18081706
Chicago/Turabian StyleWiśniewska, Anna, Rafał Olszanecki, Justyna Totoń-Żurańska, Katarzyna Kuś, Aneta Stachowicz, Maciej Suski, Anna Gębska, Mariusz Gajda, Jacek Jawień, and Ryszard Korbut. 2017. "Anti-Atherosclerotic Action of Agmatine in ApoE-Knockout Mice" International Journal of Molecular Sciences 18, no. 8: 1706. https://doi.org/10.3390/ijms18081706
APA StyleWiśniewska, A., Olszanecki, R., Totoń-Żurańska, J., Kuś, K., Stachowicz, A., Suski, M., Gębska, A., Gajda, M., Jawień, J., & Korbut, R. (2017). Anti-Atherosclerotic Action of Agmatine in ApoE-Knockout Mice. International Journal of Molecular Sciences, 18(8), 1706. https://doi.org/10.3390/ijms18081706