In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Fipronil Specific ssDNA MRE
2.2. Affinity and Specifcity of Fipronil-Specific ssDNA MRE in Selection Buffer and Buffered River Water
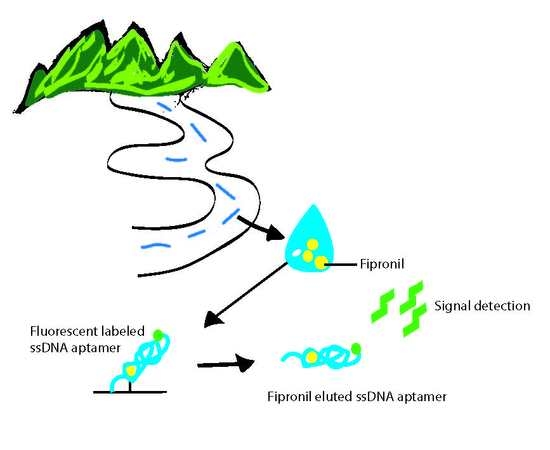
2.3. Biosensing Application of Fipronil-Specific MRE in River Water
3. Materials and Methods
3.1. In Vitro Selection for Fipronil-Specific MREs
3.2. Cloning and Sequencing of the Fipronil-Binding MREs
3.3. Fipronil-Binding MREs Binding Assays in Selection Buffer
3.4. Fipronil Cross-Binding Assay in River Water
3.5. Fipronil Detection Assay in River Water
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MRE | Molecular Recognition Element |
| BSA | Bovine Serum Albumin |
| IS | Immobilizing Substrate |
| IT | Immobilized Target |
| LOD | Limit of Detection |
References
- U.S. Environmental Protection Agency, Office of Prevention. New-Pesticide Fact. Sheet—Fipronil; Pesticides and Toxic Substances, Office of Pesticide Programs, U.S. Government Printing Office: Washington, DC, USA, 1996; pp. 1–10.
- Caboni, P.; Sammelson, R.E.; Casida, J.E. Phenylpyrazole insecticide photochemistry, metabolism, and gabaergic action: Ethiprole compared with fipronil. J. Agric. Food Chem. 2003, 51, 7055–7061. [Google Scholar] [CrossRef] [PubMed]
- Stone, W.W.; Gilliom, R.J.; Ryberg, K.R. Pesticides in U.S. Streams and rivers: Occurrence and trends during 1992–2011. Environ. Sci. Technol. 2014, 48, 11025–11030. [Google Scholar] [CrossRef] [PubMed]
- Moran, K.D. Urban Use of the Insecticide Fipronil—Water Quality Implications; TDC Environmental: San Mateo, CA, USA, 2007; pp. 1–14. [Google Scholar]
- Simon-Delso, N.; Amaral-Rogers, V.; Belzunces, L.P.; Bonmatin, J.M.; Chagnon, M.; Downs, C.; Furlan, L.; Gibbons, D.W.; Giorio, C.; Girolami, V.; et al. Systemic insecticides (neonicotinoids and fipronil): Trends, uses, mode of action and metabolites. Environ. Sci. Pollut. Res. Int. 2015, 22, 5–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquatic Life Benchmarks and Ecological Risk Assessments for Registered Pesticides. Available online: https://www.epa.gov/pesticide-science-and-assessing-pesticide-risks/aquatic-life-benchmarks-and-ecological-risk#benchmarks (accessed on 30 October 2017).
- Chau, N.D.; Sebesvari, Z.; Amelung, W.; Renaud, F.G. Pesticide pollution of multiple drinking water sources in the mekong delta, vietnam: Evidence from two provinces. Environ. Sci. Pollut. Res. Int. 2015, 22, 9042–9058. [Google Scholar] [CrossRef] [PubMed]
- Weston, D.P.; Lydy, M.J. Toxicity of the insecticide fipronil and its degradates to benthic macroinvertebrates of urban streams. Environ. Sci. Technol. 2014, 48, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Bonmatin, J.M.; Giorio, C.; Girolami, V.; Goulson, D.; Kreutzweiser, D.P.; Krupke, C.; Liess, M.; Long, E.; Marzaro, M.; Mitchell, E.A.; et al. Environmental fate and exposure; neonicotinoids and fipronil. Environ. Sci. Pollut. Res. Int. 2015, 22, 35–67. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency, Healthy Effects Division. Fipronil: Third Reevaluation—Report of the Hazard Identification Assessment Review Committee; U.S. Government Printing Office: Washington, DC, USA, 2000; pp. 1–24.
- Li, P.; Akk, G. The insecticide fipronil and its metabolite fipronil sulphone inhibit the rat α1β2γ2l GABAA receptor. Br. J. Pharmacol. 2008, 155, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Van der Sluijs, J.P.; Amaral-Rogers, V.; Belzunces, L.P.; van Bijleveld Lexmond, M.F.; Bonmatin, J.M.; Chagnon, M.; Downs, C.A.; Furlan, L.; Gibbons, D.W.; Giorio, C.; et al. Conclusions of the worldwide integrated assessment on the risks of neonicotinoids and fipronil to biodiversity and ecosystem functioning. Environ. Sci. Pollut. Res. Int. 2015, 22, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Bichon, E.; Richard, C.A.; Le Bizec, B. Development and validation of a method for fipronil residue determination in ovine plasma using 96-well plate solid-phase extraction and gas chromatography-tandem mass spectrometry. J. Chromatogr. A 2008, 1201, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Duhan, A.; Kumari, B.; Duhan, S. Determination of residues of fipronil and its metabolites in cauliflower by using gas chromatography-tandem mass spectrometry. Bull. Environ. Contam. Toxicol. 2015, 94, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, J.J.; Bernal, J.L.; del Nozal, M.J.; Martin, M.T.; Mayo, R. Sample preparation methods to analyze fipronil in honey by gas chromatography with electron-capture and mass spectrometric detection. J. Chromatogr. A 2008, 1187, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, M.Z.; Puel, S.; Toutain, P.L.; Viguie, C. Quantification of fipronil and its metabolite fipronil sulfone in rat plasma over a wide range of concentrations by LC/UV/MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2010, 878, 1934–1938. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yan, C.; Dong, J.; Yu, X.; Xu, D. Poly- and monoclonal antibody-based elisas for fipronil. J. Agric. Food Chem. 2007, 55, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, F.; Senarathna, L.; Percy, A.; Abeyewardene, M.; Eaglesham, G.; Cheng, R.; Azher, S.; Hittarage, A.; Dissanayake, W.; Sheriff, M.H.; et al. Acute human self-poisoning with the N-phenylpyrazole insecticide fipronil—A GABAA-gated chloride channel blocker. J. Toxicol. Clin. Toxicol. 2004, 42, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Vasylieva, N.; Ahn, K.C.; Barnych, B.; Gee, S.J.; Hammock, B.D. Development of an immunoassay for the detection of the phenylpyrazole insecticide fipronil. Environ. Sci. Technol. 2015, 49, 10038–10047. [Google Scholar] [CrossRef] [PubMed]
- Bordeaux, J.; Welsh, A.; Agarwal, S.; Killiam, E.; Baquero, M.; Hanna, J.; Anagnostou, V.; Rimm, D. Antibody validation. Biotechniques 2010, 48, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Mairal, T.; Ozalp, V.C.; Lozano Sanchez, P.; Mir, M.; Katakis, I.; O’Sullivan, C.K. Aptamers: Molecular tools for analytical applications. Anal. Bioanal. Chem. 2008, 390, 989–1007. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Yancey, K.; Battistella, L.; Williams, R.M.; Hickey, K.M.; Bostick, C.D.; Gannett, P.M.; Sooter, L.J. Selection of single-stranded DNA molecular recognition elements against exotoxin a using a novel decoy-selex method and sensitive detection of exotoxin a in human serum. BioMed Res. Int. 2015, 2015, 417641. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Kulick, A.R.; Yedlapalli, S.; Battistella, L.; Hajiran, C.J.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element specific for bromacil. J. Nucleic Acids 2014, 2014, 102968. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Crihfield, C.L.; Gattu, S.; Holland, L.A.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element against atrazine. Int. J. Mol. Sci. 2014, 15, 14332–14347. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Maher, E.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element for the pesticide malathion. Comb. Chem. High Throughput Screen. 2014, 17, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Connell, G.J.; Illangesekare, M.; Yarus, M. Three small ribooligonucleotides with specific arginine sites. Biochemistry 1993, 32, 5497–5502. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Khrapov, M.; Shaw, C.A. The scene of a frozen accident. RNA 2000, 6, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.; Hesselberth, J.R.; Ellington, A.D. Computational selection of nucleic acid biosensors via a slip structure model. Biosens. Bioelectron. 2007, 22, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Changayil, S.; Majerfeld, I.; Yarus, M. Selection of the simplest RNA that binds isoleucine. RNA 2003, 9, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Majerfeld, I.; Yarus, M. Isoleucine: RNA sites with associated coding sequences. RNA 1998, 4, 471–478. [Google Scholar] [PubMed]
- Alsager, O.A.; Kumar, S.; Willmott, G.R.; McNatty, K.P.; Hodgkiss, J.M. Small molecule detection in solution via the size contraction response of aptamer functionalized nanoparticles. Biosens. Bioelectron. 2014, 57, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Mehta, J.; Rouah-Martin, E.; van Dorst, B.; Maes, B.; Herrebout, W.; Scippo, M.L.; Dardenne, F.; Blust, R.; Robbens, J. Selection and characterization of PCB-binding DNA aptamers. Anal. Chem. 2012, 84, 1669–1676. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Kwon, Y.S.; Kim, J.H.; Gu, M.B. Multiple go-selex for efficient screening of flexible aptamers. Chem. Commun. 2014, 50, 10513–10516. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Battistella, L.; Salva, A.D.; Williams, R.M.; Sooter, L.J. In vitro selection of single-stranded DNA molecular recognition elements against S. aureus α toxin and sensitive detection in human serum. Int. J. Mol. Sci. 2015, 16, 2794–2809. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Maher, E.; Williams, R.M.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element against clostridium difficile toxin B and sensitive detection in human fecal matter. J. Nucleic Acids 2015, 2015, 808495. [Google Scholar] [CrossRef] [PubMed]
- Department of Environmental Protection, Commonwealth of Pennsylvania. 2012–2013 Susquehanna River Sampling and Assessment Report; Commonwealth of Pennsylvania: Harrisburg, PA, USA, 2014; pp. 1–147.
- Miyakawa, S.; Nomura, Y.; Sakamoto, T.; Yamaguchi, Y.; Kato, K.; Yamazaki, S.; Nakamura, Y. Structural and molecular basis for hyperspecificity of RNA aptamer to human immunoglobulin G. RNA 2008, 14, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Tingle, C.C.; Rother, J.A.; Dewhurst, C.F.; Lauer, S.; King, W.J. Fipronil: Environmental fate, ecotoxicology, and human health concerns. Rev. Environ. Contam. Toxicol. 2003, 176, 1–66. [Google Scholar] [PubMed]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29, S49–S52. [Google Scholar] [PubMed]
- Miyake, S.; Uchigashima, M.; Kadowaki, A. Kit for Measurement of Termite Insecticide Active Ingredient by Immunoassay Method. U.S. Patent 8,323,904 B2, 4 December 2012. [Google Scholar]
- Fan, L.; Zhao, G.; Shi, H.; Liu, M.; Li, Z. A highly selective electrochemical impedance spectroscopy-based aptasensor for sensitive detection of acetamiprid. Biosens. Bioelectron. 2013, 43, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, W.; Ma, W.; Liu, L.; Ma, W.; Zhao, Y.; Zhu, Y.; Xu, L.; Kuang, H.; Xu, C. Fluorescent strip sensor for rapid determination of toxins. Chem. Commun. 2011, 47, 1574–1576. [Google Scholar] [CrossRef] [PubMed]
- Barthelmebs, L.; Jonca, J.; Hayat, A.; Prieto-Simon, B.; Marty, J.-L. Enzyme-linked aptamer assays (ELAAs), based on a competition format for a rapid and sensitive detection of ochratoxin a in wine. Food Control 2011, 22, 737–743. [Google Scholar] [CrossRef]
- Madianos, L.; Tsekenis, G.; Skotadis, E.; Patsiouras, L.; Tsoukalas, D. A highly sensitive impedimetric aptasensor for the selective detection of acetamiprid and atrazine based on microwires formed by platinum nanoparticles. Biosens. Bioelectron. 2018, 101, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- De-Los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Modified-RNA aptamer-based sensor for competitive impedimetric assay of neomycin B. J. Am. Chem. Soc. 2007, 129, 3808–3809. [Google Scholar] [CrossRef] [PubMed]






| Round | Positive Selection | Time | Negative Selection | Time |
|---|---|---|---|---|
| 1 | Immobilized Target (IT) | 48 h | Immobilizing substrate (IS) | 24 h |
| 2 | IT | 22 h | Immobilized Negative Target (INT) Fipronil Sulfide | 24 h |
| 3 | IT | 16 h | INT Fipronil Desulfinyl | 24 h |
| 4 | IT | 11 h | INT Fipronil Sulfone | 24 h |
| 5 | IT | 6.5 h | INT Ethiprole | 24 h |
| 6 | IT | 3 h | IT/ 1 mM of BSA Free Elution (FE) | 3 h/ 24 h |
| 7 | IT/ Competitive Elution with 500 µM free Fipronil | 3 h/ 1 h | IT/ 1 µM of Atrazine FE/1 µM of Propanil FE | 3 h/ 24 h/ 24 h |
| 8 | IT/ Competitive Elution with 500 µM free Fipronil | 1 h/ 30 min | IT/ 1 µM of Fipronil Sulfone FE/ 1 µM of Fipronil Desulfinyl FE | 3 h/ 24 h/ 24 h |
| 9 | IT/ Competitive Elution with 100 µM free Fipronil | 15 min/ 5 min | IT/ 1 µM of Fipronil Sulfide FE | 15 min/ 15 min |
| 10 | IT/ Competitive Elution with 10 µM free Fipronil | 1 min/ 1 min | IT/ 1 µM of Ethiprole Free Elution | 1 min/ 1 min |
| 11 | IT/ Competitive Elution with 1 µM free Fipronil | Immediate/ Immediate | IT/ 1 mM of BSA FE/ 1 µM of Malathion FE | 1 min/ 6 h/ 30 min |
| 12 | IT/ Competitive Elution with 100 nM free Fipronil | Immediate/ Immediate | - |
| Target | Normalized Average Fluorescence | Standard Deviation | p-Value | Selective Ratio |
|---|---|---|---|---|
| Fipronil | 2.413 | 0.194 | - | - |
| Fipronil Sulfone | 2.248 | 0.388 | 0.279 | 1.07 |
| Fipronil Sulfide | 2.726 | 0.460 | 0.169 | 0.89 |
| Fipronil Desulfinyl | 2.088 | 0.623 | 0.219 | 1.16 |
| Ethiprole | 2.185 | 0.217 | 0.124 | 1.10 |
| Atrazine | 1.963 | 0.227 | 0.030 | 1.23 |
| Propanil | 1.587 | 0.243 | 0.005 | 1.52 |
| Malathion | 2.048 | 0.184 | 0.039 | 1.18 |
| BSA | 8.687 | 0.360 | 5.99 × 10−6 | 0.28 |
| Selection Buffer (No Targets) | 0.247 | 0.214 | 1.02 × 10−4 | 9.74 |
| Target | Normalized Average Fluorescence | Standard Deviation | p-Value | Selective Ratio |
|---|---|---|---|---|
| Fipronil | 3.863 | 0.090 | - | - |
| Fipronil Sulfone | 2.863 | 0.180 | 4.98 × 10−4 | 1.34 |
| Fipronil Sulfide | 4.333 | 0.944 | 2.19 × 10−1 | 0.89 |
| Fipronil Desulfinyl | 2.902 | 0.245 | 15.5 × 10−3 | 1.33 |
| Ethiprole | 3.157 | 0.148 | 1.06 × 10−3 | 1.22 |
| Atrazine | 0.804 | 0.302 | 3.66 × 10−5 | 4.80 |
| Propanil | 0.764 | 0.509 | 2.44 × 10−4 | 5.05 |
| Malathion | 0.725 | 0.355 | 5.98 × 10−5 | 5.32 |
| BSA | 20.41 | 1.118 | 6.95 × 10−6 | 0.19 |
| River water/Buffer Mix (No Targets) | −0.549 | 1.779 | 6.37 × 10−3 | −7.035 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, K.L.; Sooter, L.J. In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water. Int. J. Mol. Sci. 2018, 19, 85. https://doi.org/10.3390/ijms19010085
Hong KL, Sooter LJ. In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water. International Journal of Molecular Sciences. 2018; 19(1):85. https://doi.org/10.3390/ijms19010085
Chicago/Turabian StyleHong, Ka L., and Letha J. Sooter. 2018. "In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water" International Journal of Molecular Sciences 19, no. 1: 85. https://doi.org/10.3390/ijms19010085
APA StyleHong, K. L., & Sooter, L. J. (2018). In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water. International Journal of Molecular Sciences, 19(1), 85. https://doi.org/10.3390/ijms19010085