Comparative Transcriptomic and Lipidomic Analyses of Human Male and Female Meibomian Glands Reveal Common Signature Genes of Meibogenesis
Abstract
:1. Introduction
2. Results
2.1. Comparative Lipidomic Analyses of Human Male and Female Meibum
2.2. Untargeted Transcriptomic Analyses of Male and Female Tarsal Plates
2.3. Comparative Transcriptomic Analyses of Meibomian Glands and Tarsal Plate Connective Tissues
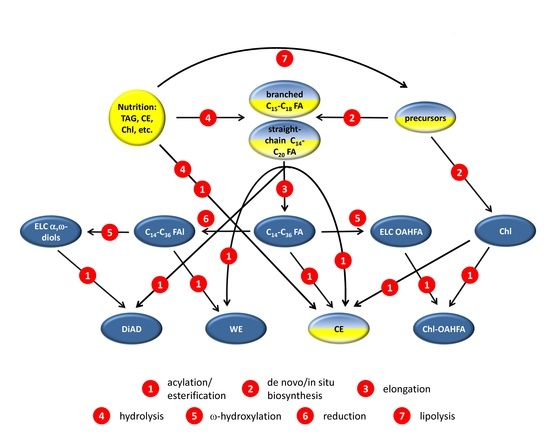
2.4. Targeted Transcriptomic Analyses of Male and Female Tarsal Plates
3. Discussion
4. Materials and Methods
4.1. Collection and Preparation of Study Samples
4.2. Analytical Procedures
4.3. Data Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | Atmospheric pressure chemical ionization |
| CE | Cholesteryl ester(s) |
| Ch | Free cholesterol |
| Chl-OAHFA | Cholesteryl ester(s) of (O)-acylated omega-hydroxy fatty acid(s) |
| CT | Connective tissue |
| DiAD | Diacylated diol(s) |
| ELCFA | Extremely long chain fatty acid(s) |
| FA | Fatty acid(s) |
| FAl | Fatty alcohol(s) |
| GEP | Gene expression profile(s) |
| LFC | Linear fold change |
| MG | Meibomian gland(s) |
| MGD | Meibomian gland dysfunction |
| MS | Mass spectrometry |
| NIM | Negative ion mode |
| OAHFA | (O)-acylated omega-hydroxy fatty acid(s) |
| PCA | Principal component analysis |
| PIM | positive ion mode |
| RP | Reversed phase |
| RT | Retention time(s) |
| TAG | Triacylglycerol(s) |
| TIC | Total ion chromatogram(s) |
| TOF | Time-of-Flight |
| TP | Tarsal plate(s) |
| UPLC | Ultra-high performance liquid chromatography |
| VLCFA | Very long chain fatty acid(s) |
| WE | Wax ester(s) |
References
- Meibom, H. De Vasis Palpebrarum Novis Epistola; Henningi Mulleri: Helmstadt, Germany, 1666. [Google Scholar]
- Nicolaides, N.; Kaitaranta, J.K.; Rawdah, T.N.; Macy, J.I.; Boswell, F.M., 3rd; Smith, R.E. Meibomian gland studies: Comparison of steer and human lipids. Investig. Ophthalmol. Vis. Sci. 1981, 20, 522–536. [Google Scholar] [PubMed]
- Nelson, J.D.; Shimazaki, J.; Benitez-del-Castillo, J.M.; Craig, J.P.; McCulley, J.P.; Den, S.; Foulks, G.N. The international workshop on meibomian gland dysfunction: Report of the definition and classification subcommittee. Investig. Ophthalmol. Vis. Sci. 2011, 52, 1930–1937. [Google Scholar] [CrossRef] [PubMed]
- Souchier, M.; Joffre, C.; Gregoire, S.; Bretillon, L.; Muselier, A.; Acar, N.; Beynat, J.; Bron, A.; D’Athis, P.; Creuzot-Garcher, C. Changes in meibomian fatty acids and clinical signs in patients with meibomian gland dysfunction after minocycline treatment. Br. J. Ophthalmol. 2008, 92, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Joffre, C.; Souchier, M.; Gregoire, S.; Viau, S.; Bretillon, L.; Acar, N.; Bron, A.M.; Creuzot-Garcher, C. Differences in meibomian fatty acid composition in patients with meibomian gland dysfunction and aqueous-deficient dry eye. Br. J. Ophthalmol. 2008, 92, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Arita, R.; Mori, N.; Shirakawa, R.; Asai, K.; Imanaka, T.; Fukano, Y.; Nakamura, M. Linoleic acid content of human meibum is associated with telangiectasia and plugging of gland orifices in meibomian gland dysfunction. Exp. Eye Res. 2016, 145, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Matossian, C.; McDonald, M.; Donaldson, K.E.; Nichols, K.K.; MacIver, S.; Gupta, P.K. Dry Eye Disease: Consideration for Women’s Health. J. Womens Health (Larchmt). 2019, 28, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Viso, E.; Rodriguez-Ares, M.T.; Abelenda, D.; Oubina, B.; Gude, F. Prevalence of asymptomatic and symptomatic meibomian gland dysfunction in the general population of Spain. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2601–2606. [Google Scholar] [CrossRef]
- Suzuki, T.; Fujiwara, S.; Kinoshita, S.; Butovich, I.A. Cyclic Change of Fatty Acid Composition in Meibum During the Menstrual Cycle. Investig. Ophthalmol. Vis. Sci. 2019, 60, 1724–1733. [Google Scholar] [CrossRef]
- Butovich, I.A.; McMahon, A.; Wojtowicz, J.C.; Lin, F.; Mancini, R.; Itani, K. Dissecting lipid metabolism in meibomian glands of humans and mice: An integrative study reveals a network of metabolic reactions not duplicated in other tissues. Biochim. Biophys. Acta 2016, 1861, 538–553. [Google Scholar] [CrossRef] [Green Version]
- Butovich, I.A. Meibomian glands, meibum, and meibogenesis. Exp. Eye Res. 2017, 163, 2–16. [Google Scholar] [CrossRef]
- Sullivan, B.D.; Evans, J.E.; Krenzer, K.L.; Reza Dana, M.; Sullivan, D.A. Impact of antiandrogen treatment on the fatty acid profile of neutral lipids in human meibomian gland secretions. J. Clin. Endocrinol. Metab. 2000, 85, 4866–4873. [Google Scholar] [CrossRef] [PubMed]
- Wickham, L.A.; Gao, J.; Toda, I.; Rocha, E.M.; Ono, M.; Sullivan, D.A. Identification of androgen, estrogen and progesterone receptor mRNAs in the eye. Acta Ophthalmol. Scand. 2000, 78, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, D.A.; Jensen, R.V.; Suzuki, T.; Richards, S.M. Do sex steroids exert sex-specific and/or opposite effects on gene expression in lacrimal and meibomian glands? Mol. Vis. 2009, 15, 1553–1572. [Google Scholar] [PubMed]
- Sullivan, B.D.; Evans, J.E.; Cermak, J.M.; Krenzer, K.L.; Dana, M.R.; Sullivan, D.A. Complete androgen insensitivity syndrome: Effect on human meibomian gland secretions. Arch. Ophthalmol. 2002, 120, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.M.; Jensen, R.V.; Liu, M.; Sullivan, B.D.; Lombardi, M.J.; Rowley, P.; Schirra, F.; Treister, N.S.; Suzuki, T.; Steagall, R.J.; et al. Influence of sex on gene expression in the mouse lacrimal gland. Exp. Eye Res. 2006, 82, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Butovich, I.A. Tear film lipids. Exp. Eye Res. 2013, 117, 4–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butovich, I.A. Lipidomics of human Meibomian gland secretions: Chemistry, biophysics, and physiological role of Meibomian lipids. Prog. Lipid Res. 2011, 50, 278–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butovich, I.A. The Meibomian puzzle: Combining pieces together. Prog. Retin Eye Res. 2009, 28, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Butovich, I.A.; Lu, H.; McMahon, A.; Eule, J.C. Toward an animal model of the human tear film: Biochemical comparison of the mouse, canine, rabbit, and human meibomian lipidomes. Investig. Ophthalmol. Vis. Sci. 2012, 53, 6881–6896. [Google Scholar] [CrossRef] [PubMed]
- Butovich, I.A.; McMahon, A.; Wojtowicz, J.C.; Bhat, N.; Wilkerson, A. Effects of sex (or lack thereof) on meibogenesis in mice (Mus musculus): Comparative evaluation of lipidomes and transcriptomes of male and female tarsal plates. Ocul. Surf. 2019. [Google Scholar] [CrossRef] [PubMed]
- Butovich, I.A.; Wilkerson, A.; Bhat, N.; McMahon, A.; Yuksel, S. On the pivotal role of Elovl3/ELOVL3 in meibogenesis and ocular physiology of mice. FASEB J. 2019, fj201900725R. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, D.A.; Rocha, E.M.; Ullman, M.D.; Krenzer, K.L.; Gao, J.; Toda, I.; Dana, M.R.; Bazzinotti, D.; da Silveira, L.A.; Wickham, L.A. Androgen regulation of the meibomian gland. Adv. Exp. Med. Biol. 1998, 438, 327–331. [Google Scholar] [PubMed]
- Khandelwal, P.; Liu, S.; Sullivan, D.A. Androgen regulation of gene expression in human meibomian gland and conjunctival epithelial cells. Mol. Vis. 2012, 18, 1055–1067. [Google Scholar] [PubMed]
- Suzuki, T.; Schirra, F.; Richards, S.M.; Jensen, R.V.; Sullivan, D.A. Estrogen and progesterone control of gene expression in the mouse meibomian gland. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1797–1808. [Google Scholar] [CrossRef]
- Luu-The, V. Assessment of steroidogenesis and steroidogenic enzyme functions. J. Steroid Biochem. Mol. Biol. 2013, 137, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Butovich, I.A.; Uchiyama, E.; McCulley, J.P. Lipids of human meibum: Mass-spectrometric analysis and structural elucidation. J. Lipid Res. 2007, 48, 2220–2235. [Google Scholar] [CrossRef]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef]
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef]
- Sassa, T.; Tadaki, M.; Kiyonari, H.; Kihara, A. Very long-chain tear film lipids produced by fatty acid elongase ELOVL1 prevent dry eye disease in mice. FASEB J. 2018, 32, 2966–2978. [Google Scholar] [CrossRef]
- Butovich, I.A.; Arciniega, J.C.; Lu, H.; Molai, M. Evaluation and quantitation of intact wax esters of human meibum by gas-liquid chromatography-ion trap mass spectrometry. Investig. Ophthalmol. Vis. Sci. 2012, 53, 3766–3781. [Google Scholar] [CrossRef] [PubMed]
- Heid, H.; Rickelt, S.; Zimbelmann, R.; Winter, S.; Schumacher, H.; Dorflinger, Y. Lipid droplets, perilipins and cytokeratins--unravelled liaisons in epithelium-derived cells. PLoS ONE 2013, 8, e63061. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sullivan, B.D.; Darabad, R.R.; Liu, S.; Kam, W.R.; Sullivan, D.A. Are BALB/c Mice Relevant Models for Understanding Sex-Related Differences in Gene Expression in the Human Meibomian Gland? Cornea 2019. [Google Scholar] [CrossRef] [PubMed]
- Arita, R.; Mori, N.; Shirakawa, R.; Asai, K.; Imanaka, T.; Fukano, Y.; Nakamura, M.; Amano, S. Meibum Color and Free Fatty Acid Composition in Patients With Meibomian Gland Dysfunction. Investig. Ophthalmol. Vis. Sci. 2015, 56, 4403–4412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, H.S.; Parfitt, G.J.; Brown, D.J.; Jester, J.V. Meibocyte differentiation and renewal: Insights into novel mechanisms of meibomian gland dysfunction (MGD). Exp. Eye Res. 2017, 163, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Jester, J.V.; Potma, E.; Brown, D.J. PPARgamma Regulates Mouse Meibocyte Differentiation and Lipid Synthesis. Ocul. Surf. 2016, 14, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Hampel, U.; Schroder, A.; Mitchell, T.; Brown, S.; Snikeris, P.; Garreis, F.; Kunnen, C.; Willcox, M.; Paulsen, F. Serum-induced keratinization processes in an immortalized human meibomian gland epithelial cell line. PLoS ONE 2015, 10, e0128096. [Google Scholar] [CrossRef] [PubMed]
- Bekerman, I.; Gottlieb, P.; Vaiman, M. Variations in eyeball diameters of the healthy adults. J. Ophthalmol. 2014, 2014, 503645. [Google Scholar] [CrossRef] [PubMed]
- Butovich, I.A. Lipidomic analysis of human meibum using HPLC-MSn. Methods Mol. Biol. 2009, 579, 221–246. [Google Scholar]








| Gene | Females, Mean Bi-Weight Signal, Log2 | Females, Standard Deviation, Log2 | Males, Mean Bi-Weight Signal, Log2 | Males, Standard Deviation, Log2 | Significance, p (α = 0.05) |
|---|---|---|---|---|---|
| AADACL4 | 13.48 | 1.69 | 13.82 | 0.72 | 0.690 |
| ABHD5 | 12.94 | 1.44 | 12.49 | 0.67 | 0.544 |
| ACADVL | 10.06 | 0.73 | 11.59 | 0.18 | 0.002 * |
| ACAT2 | 15.47 | 1.31 | 15.05 | 1.13 | 0.602 |
| ACSL1 | 13.3 | 1.11 | 13.24 | 0.7 | 0.921 |
| ACSL3 | 16.27 | 1.03 | 16.24 | 0.75 | 0.959 |
| ACSL5 | 11.56 | 1.13 | 11.65 | 0.94 | 0.894 |
| APOD | 13.24 | 1.29 | 14.24 | 0.6 | 0.155 |
| AWAT1 | 15.01 | 1.96 | 13.28 | 1.46 | 0.152 |
| AWAT2 | 16.7 | 1.07 | 16.54 | 0.55 | 0.774 |
| CES1 | 13.35 | 0.89 | 14.39 | 1.64 | 0.48 |
| CYP4F22 | 13.38 | 1.12 | 12.97 | 0.6 | 0.491 |
| CYP4X1 | 12.2 | 0.45 | 13.42 | 0.94 | 0.031 * |
| CYP51A1 | 13.42 | 1.33 | 13.15 | 1.11 | 0.736 |
| DBT | 11.18 | 0.81 | 11.25 | 0.79 | 0.893 |
| DGAT2 | 13.96 | 1.28 | 13.01 | 1.17 | 0.255 |
| DHCR24 | 17.26 | 0.66 | 16.79 | 0.67 | 0.296 |
| DHCR7 | 14.7 | 1.14 | 13.54 | 0.73 | 0.092 |
| ECHDC1 | 12.53 | 0.81 | 12.2 | 0.58 | 0.480 |
| EGFR | 12.27 | 0.38 | 12.4 | 0.67 | 0.716 |
| ELOVL1 | 11.98 | 1.07 | 11.74 | 0.75 | 0.692 |
| ELOVL2 | 5.68 | 0.76 | 5.18 | 0.29 | 0.207 |
| ELOVL3 | 14.58 | 1.11 | 14.79 | 1 | 0.761 |
| ELOVL4 | 18.71 | 0.9 | 17.62 | 0.86 | 0.086 |
| ELOVL5 | 9.79 | 1.15 | 9.21 | 0.62 | 0.35 |
| ELOVL6 | 14.41 | 0.85 | 14.36 | 1 | 0.934 |
| ELOVL7 | 9.04 | 0.75 | 8.55 | 0.38 | 0.229 |
| FAR2 | 18.02 | 0.83 | 17.08 | 0.81 | 0.107 |
| FASN | 11.86 | 1 | 11.07 | 0.52 | 0.156 |
| GPAM | 11.15 | 1.11 | 10.61 | 0.73 | 0.390 |
| HMGCR | 17.12 | 1.34 | 16.92 | 0.84 | 0.785 |
| HMGCS1 | 16.79 | 0.75 | 16.47 | 0.95 | 0.571 |
| HSD11B1 | 11.45 | 1.45 | 11.46 | 1.06 | 0.99 |
| HSD17B12 | 13.85 | 0.63 | 13.04 | 1.24 | 0.229 |
| HSD17B2 | 15.41 | 1.15 | 15.36 | 0.5 | 0.931 |
| HSD17B4 | 11.65 | 0.29 | 11.97 | 0.22 | 0.085 |
| HSDL2 | 12.46 | 0.51 | 12.15 | 0.6 | 0.404 |
| IFITM2 | 14.12 | 1.75 | 12.33 | 0.63 | 0.064 |
| MSMO1 | 16.61 | 0.8 | 16.23 | 0.98 | 0.521 |
| PEBP1 | 13.75 | 0.7 | 13.77 | 0.31 | 0.955 |
| SCARB2 | 11.36 | 0.7 | 11.43 | 0.38 | 0.849 |
| SCD | 19.41 | 0.4 | 19.24 | 0.39 | 0.515 |
| SLC12A2 | 13.97 | 2.01 | 12.62 | 1.46 | 0.259 |
| SLC25A6 | 13.44 | 0.33 | 13.29 | 0.48 | 0.581 |
| SLC31A1 | 14.46 | 1.36 | 13.03 | 0.54 | 0.06 |
| SLC38A2 | 14.95 | 0.71 | 15.34 | 0.56 | 0.363 |
| SLCO4C1 | 13.87 | 1.41 | 11.77 | 0.97 | 0.025 * |
| SOAT1 | 17.2 | 0.75 | 17.47 | 0.9 | 0.620 |
| SOAT2 | 5.13 | 0.25 | 4.90 | 0.39 | 0.299 |
| SQLE | 13.49 | 1.4 | 12.9 | 0.95 | 0.458 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butovich, I.A.; Bhat, N.; Wojtowicz, J.C. Comparative Transcriptomic and Lipidomic Analyses of Human Male and Female Meibomian Glands Reveal Common Signature Genes of Meibogenesis. Int. J. Mol. Sci. 2019, 20, 4539. https://doi.org/10.3390/ijms20184539
Butovich IA, Bhat N, Wojtowicz JC. Comparative Transcriptomic and Lipidomic Analyses of Human Male and Female Meibomian Glands Reveal Common Signature Genes of Meibogenesis. International Journal of Molecular Sciences. 2019; 20(18):4539. https://doi.org/10.3390/ijms20184539
Chicago/Turabian StyleButovich, Igor A., Nita Bhat, and Jadwiga C. Wojtowicz. 2019. "Comparative Transcriptomic and Lipidomic Analyses of Human Male and Female Meibomian Glands Reveal Common Signature Genes of Meibogenesis" International Journal of Molecular Sciences 20, no. 18: 4539. https://doi.org/10.3390/ijms20184539
APA StyleButovich, I. A., Bhat, N., & Wojtowicz, J. C. (2019). Comparative Transcriptomic and Lipidomic Analyses of Human Male and Female Meibomian Glands Reveal Common Signature Genes of Meibogenesis. International Journal of Molecular Sciences, 20(18), 4539. https://doi.org/10.3390/ijms20184539