Effects of Single and Double Mutants in Human Glucose-6-Phosphate Dehydrogenase Variants Present in the Mexican Population: Biochemical and Structural Analysis
Abstract
:1. Introduction
2. Results and Discussion
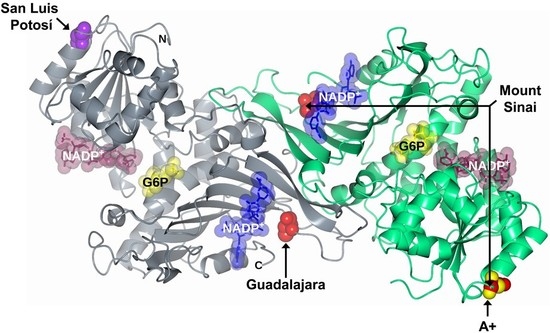
2.1. In Silico Mutagenesis and Modeling
2.2. Construction Expression and Purification of Recombinant G6PD Variants
2.3. Determination of Steady-State Kinetic Parameters
2.4. Circular Dichroism (DC) Analysis
2.5. Structural Analysis by Intrinsic and 8-Anilinonaphthalene-1-Sulfonate (ANS) Binding Assays
2.6. Evaluation of Stability of the Variants
2.6.1. Thermal Stability Analysis of Recombinant G6PD Variants
2.6.2. Thermal Inactivation Assays
2.6.3. Stability Analysis of G6PD Variants in the Presence of Guanidine Hydrochloride
2.6.4. Susceptibility to Trypsin Digestion Assays
3. Materials and Methods
3.1. In Silico Mutagenesis and Modeling
3.2. Protein Stability Prediction in 3D
3.3. Construction of Recombinant G6PD Variants by Site-Directed Mutagenesis
3.4. Expression and Purification of G6PD Recombinant Variants
3.5. Determination of Steady-State Kinetic Parameters
3.6. Circular Dichroism (DC) Analysis
3.7. Structural Analysis by Intrinsic and 8-Anilinonaphthalene-1-Sulfonate (ANS) Binding Assays
3.8. Evaluation of Stability of the Variants
3.8.1. Thermal Stability Analysis of Recombinant G6PD Variants
3.8.2. Thermal Inactivation Assays
3.8.3. Stability Analysis of G6PD Variants in the Presence of Guanidine Hydrochloride
3.8.4. Thermal Stability Analysis of Recombinant G6PD Variants
3.8.5. Susceptibility to Trypsin Digestion Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eggleston, L.V.; Krebs, H.A. Regulation of the pentose phosphate cycle. Biochem. J. 1974, 138, 425–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luzzatto, L.; Nannelli, C.; Notaro, R. Glucose-6-phosphate dehydrogenase deficiency. Hematol. Oncol. Clin. N. Am. 2016, 30, 373–393. [Google Scholar] [CrossRef] [PubMed]
- Mason, P.J.; Bautista, J.M.; Gilsanz, F. G6PD deficiency: The genotype-phenotype association. Blood Rev. 2007, 21, 267–283. [Google Scholar] [CrossRef] [PubMed]
- DePina, A.J.; Pires, C.M.; Andrade, A.J.B.; Dia, A.K.; Moreira, A.L.; Ferreira, M.C.M.; Correia, A.J.; Faye, O.; Seck, I.; Niang, E.H.A. The prevalence of glucose-6-phosphate dehydrogenase deficiency in the Cape Verdean population in the context of malaria elimination. PLoS ONE 2020, 15, e0229574. [Google Scholar] [CrossRef] [PubMed]
- Gampio Gueye, N.S.; Peko, S.M.; Nderu, D.; Koukouikila-Koussounda, F.; Vouvoungui, C.; Kobawila, S.C.; Velavan, T.P.; Ntoumi, F. An update on glucose-6-phosphate dehydrogenase deficiency in children from Brazzaville, Republic of Congo. Malar. J. 2019, 18, 57. [Google Scholar] [CrossRef]
- Mbanefo, E.C.; Ahmed, A.M.; Titouna, A.; Elmaraezy, A.; Trang, N.T.; Phuoc Long, N.; Hoang Anh, N.; Diem Nghi, T.; The Hung, B.; Van Hieu, M.; et al. Association of glucose-6-phosphate dehydrogenase deficiency and malaria: A systematic review and meta-analysis. Sci. Rep. 2017, 7, 45963. [Google Scholar] [CrossRef] [Green Version]
- Okafor, I.M.; Okoroiwu, H.U.; Ekechi, C.A. Hemoglobin S and Glucose-6-Phosphate Dehydrogenase Deficiency Coinheritance in AS and SS Individuals in Malaria-Endemic Region: A Study in Calabar, Nigeria. J. Glob. Infect. Dis. 2019, 11, 118–122. [Google Scholar] [CrossRef]
- Nkhoma, E.T.; Poole, C.; Vannappagari, V.; Hall, S.A.; Beutler, E. The global prevalence of glucose-6-phosphate dehydrogenase deficiency: A systematic review and meta-analysis. Blood Cells Mol. Dis. 2009, 42, 267–278. [Google Scholar] [CrossRef]
- Cappellini, M.D.; Fiorelli, G. Glucose-6-phosphate dehydrogenase deficiency. Lancet. 2008, 371, 64–74. [Google Scholar] [CrossRef]
- WHO Working Group. Glucose-6-phosphate dehydrogenase deficiency. WHO Bull. OMS. 1989, 67, 601–611. [Google Scholar]
- Gómez-Manzo, S.; Marcial-Quino, J.; Vanoye-Carlo, A.; Serrano-Posada, H.; González-Valdez, A.; Martínez-Rosas, V.; Hernández-Ochoa, B.; Sierra-Palacios, E.; Castillo-Rodríguez, R.A.; Cuevas-Cruz, M.; et al. Functional and biochemical characterization of three recombinant human Glucose-6-Phosphate Dehydrogenase mutants: Zacatecas, Vanua-Lava and Viangchan. Int. J. Mol. Sci. 2016, 17, 787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minucci, A.; Moradkhani, K.; Hwang, M.J.; Zuppi, C.; Giardina, B.; Capoluongo, E. Glucose-6-phosphate dehydrogenase (G6PD) mutations database: Review of the “old” and update of the new mutations. Blood Cells Mol. Dis. 2012, 48, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Vaca, G.; Arámbula, E.; Monsalvo, A.; Medina, C.; Nuñez, C.; Sandoval, L.; López-Guido, B. Glucose-6-phosphate dehydrogenase (G-6-PD) mutations in Mexico: Four new G-6-PD variants. Blood Cells Mol. Dis. 2003, 31, 112–120. [Google Scholar] [CrossRef]
- García-Magallanes, N.; Luque-Ortega, F.; Aguilar-Medina, E.M.; Ramos-Payán, R.; Galaviz-Hernandez, C.; Romero-Quintana, J.G.; Del pozo-Yauner, L.; Rangel-Villalobos, H.; Arámbula-Meraz, E. Glucose-6-phosphate dehydrogenase deficiency in northern Mexico and description of a novel mutation. J. Genet. 2014, 93, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Nava, E.J.; Ortega-Cuellar, D.; Serrano-Posada, H.; González-Valdez, A.; Vanoye-Carlo, A.; Hernández-Ochoa, B.; Sierra-Palacios, E.; Hernández-Pineda, J.; Rodríguez-Bustamante, E.; Arreguin-Espinosa, R.; et al. Biochemical Analysis of Two Single Mutants that Give Rise to a Polymorphic G6PD A-Double Mutant. Int. J. Mol. Sci. 2017, 18, 2244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlachos, A.; Westwood, B.; Lipton, J.M.; Beutler, E. G6PD Mount Sinai: A new severe hemolytic variant characterized by dual mutations at nucleotides 376G and 1159T (N126D). Hum. Mutat. 1998, 11, S154–S155. [Google Scholar] [CrossRef]
- Takizawa, T.; Yoneyama, Y.; Miwa, S.; Yoshida, A. A single nucleotide base transition is the basis of the common human glucose-6-phosphate dehydrogenase variant A (+). Genomics 1987, 1, 228–231. [Google Scholar] [CrossRef]
- Vaca, G.; Ibarra, B.; Romero, F.; Olivares, N.; Cantú, J.M.; Beutler, E. G-6-PD Guadalajara. A new mutant associated with chronic nonspherocytic hemolytic anemia. Hum. Genet. 1982, 61, 175–176. [Google Scholar] [CrossRef]
- Gómez-Manzo, S.; Terrón-Hernández, J.; De la Mora-De la Mora, I.; González-Valdez, A.; Marcial-Quino, J.; García-Torres, I.; Vanoye-Carlo, A.; López-Velázquez, G.; Hernández-Alcantara, G.; Oria-Hernández, J.; et al. The stability of G6PD is affected by mutations with different clinical phenotypes. Int. J. Mol. Sci. 2014, 15, 21179–21201. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.T.; Lam, V.M.; Engel, P.C. Marked decrease in specific activity contributes to disease phenotype in two human glucose-6-phosphate dehydrogenase mutants, G6PD Union and G6PD Andalus. Hum. Mutat. 2005, 26, 284–293. [Google Scholar] [CrossRef]
- Gómez-Manzo, S.; Marcial-Quino, J.; Ortega-Cuellar, D.; Serrano-Posada, H.; González-Valdez, A.; Vanoye-Carlo, A.; Hernández-Ochoa, B.; Sierra-Palacios, E.; Castillo-Villanueva, A.; Reyes-Vivas, H. Functional and biochemical analysis of glucose-6-phosphate dehydrogenase (G6PD) variants: Elucidating the molecular basis of G6PD deficiency. Catalysis 2017, 7, 135. [Google Scholar] [CrossRef] [Green Version]
- Boonyuen, U.; Chamchoy, K.; Swangsri, T.; Junkree, T.; Day, N.P.; White, N.J.; Imwong, M. A trade off between catalytic activity and protein stability determines the clinical manifestations of glucose-6-phosphate dehydrogenase (G6PD) deficiency. Int. J. Biol. Macromol. 2017, 104, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Getov, I.; Petukh, M.; Alexov, E. SAAFEC: Predicting the Effect of Single Point Mutations on Protein Folding Free Energy Using a Knowledge-Modified MM/PBSA Approach. Int. J. Mol. Sci. 2016, 17, 512. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Lam, V.M.S.; Engel, P.C. Functional properties of two mutants of human glucose 6-phosphate dehydrogenase, R393G and R393H, corresponding to the clinical variants G6PD Wisconsin and Nashville. Biochim. Biophys. Acta 2006, 1762, 767–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Manzo, S.; Marcial-Quino, J.; Vanoye-Carlo, A.; Enríquez-Flores, S.; De la Mora-De la Mora, I.; González-Valdez, A.; García-Torres, I.; Martínez-Rosas, V.; Sierra-Palacios, E.; Lazcano-Pérez, F.; et al. Mutations of glucose-6-phosphate dehydrogenase Durham, Santa-Maria and A+ variants are associated with loss functional and structural stability of the protein. Int. J. Mol. Sci. 2015, 16, 28657–28668. [Google Scholar] [CrossRef]
- Wang, X.T.; Engel, P.C. Clinical mutants of human glucose-6-phosphate dehydrogenase: Impairment of NADP+ binding affects both folding and stability. Biochim. Biophys. Acta 2009, 1792, 804–809. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of coot. Acta Crystallogr. 2010, 66, 486–501. [Google Scholar]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 2009, 77, 114–122. [Google Scholar] [CrossRef] [Green Version]
- McNicholas, S.; Potterton, E.; Wilson, K.S.; Noble, M.E.M. Presenting your structures: The CCP4mg molecular-graphics software. Acta Cryst. D. Biol. Cryst. 2011, 67, 386–394. [Google Scholar] [CrossRef] [Green Version]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Manzo, S.; Terrón-Hernández, J.; de la Mora-de la Mora, I.; García Torres, I.; López-Velázquez, G.; Reyes-Vivas, H.; Oria-Hernández, J. Cloning, expression, purification and characterization of His-tagged human glucose-6-phosphate dehydrogenase: A simplified method for protein yield. Protein J. 2013, 32, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Cortés-Morales, Y.Y.; Vanoye-Carlo, A.; Castillo-Rodríguez, R.A.; Serrano-Posada, H.; González-Valdez, A.; Ortega-Cuellar, D.; Hernández-Ochoa, B.; Moreno-Vargas, L.M.; Prada-Gracia, D.; Sierra-Palacios, E.; et al. Cloning and biochemical characterization of three glucose-6-phosphate dehydrogenase mutants presents in the Mexican population. Int. J. Biol. Macromol. 2018, 119, 926–936. [Google Scholar] [CrossRef] [PubMed]












| G6PD | Amino Acid Substitution and Class | Total Protein (mg) | Specific Activity (IU.mg−1) | Total Activity (IU) | Yield (%) |
|---|---|---|---|---|---|
| WT | - | 2.1 | 230 | 483 | 61 |
| A+ | N126D (III) | 1.8 | 114 | 205 | 43 |
| San Luis Potosi | N126Y (II) | 0.4 | 15 | 6 | 2 |
| Guadalajara | R387C (I) | 1.8 | 15 | 27 | 3 |
| Mount Sinai | N126D/R387C (I) | 1.8 | 16 | 29 | 2 |
| G6PD | Class | Amino Acid Substitution | kcat (s−1) | KmG6P (µM) | KmNADP+ (µM) | kcat/KmG6P (s−1 M−1) | kcat/KmNADP+ (s−1 M−1) |
|---|---|---|---|---|---|---|---|
| WT | - | 230 | 38.4 | 6.1 | 5.9 | 37.3 | |
| A+ | III | Asn126Asp | 114 | 56.4 | 12.9 | 2.0 | 8.7 |
| San Luis Potosi | II | Asn126Tyr | 10.4 | 43.7 | 11.9 | 1.35 | 0.28 |
| Guadalajara | I | Arg387Cys | 25.9 | 98.8 | 41.4 | 0.15 | 0.37 |
| Mount Sinai | I | Asn126Asp/ Arg387Cys | 13.9 | 27.1 | 41.6 | 0.59 | 0.38 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Rosas, V.; Juárez-Cruz, M.V.; Ramírez-Nava, E.J.; Hernández-Ochoa, B.; Morales-Luna, L.; González-Valdez, A.; Serrano-Posada, H.; Cárdenas-Rodríguez, N.; Ortiz-Ramírez, P.; Centeno-Leija, S.; et al. Effects of Single and Double Mutants in Human Glucose-6-Phosphate Dehydrogenase Variants Present in the Mexican Population: Biochemical and Structural Analysis. Int. J. Mol. Sci. 2020, 21, 2732. https://doi.org/10.3390/ijms21082732
Martínez-Rosas V, Juárez-Cruz MV, Ramírez-Nava EJ, Hernández-Ochoa B, Morales-Luna L, González-Valdez A, Serrano-Posada H, Cárdenas-Rodríguez N, Ortiz-Ramírez P, Centeno-Leija S, et al. Effects of Single and Double Mutants in Human Glucose-6-Phosphate Dehydrogenase Variants Present in the Mexican Population: Biochemical and Structural Analysis. International Journal of Molecular Sciences. 2020; 21(8):2732. https://doi.org/10.3390/ijms21082732
Chicago/Turabian StyleMartínez-Rosas, Víctor, Merit Valeria Juárez-Cruz, Edson Jiovany Ramírez-Nava, Beatriz Hernández-Ochoa, Laura Morales-Luna, Abigail González-Valdez, Hugo Serrano-Posada, Noemí Cárdenas-Rodríguez, Paulina Ortiz-Ramírez, Sara Centeno-Leija, and et al. 2020. "Effects of Single and Double Mutants in Human Glucose-6-Phosphate Dehydrogenase Variants Present in the Mexican Population: Biochemical and Structural Analysis" International Journal of Molecular Sciences 21, no. 8: 2732. https://doi.org/10.3390/ijms21082732
APA StyleMartínez-Rosas, V., Juárez-Cruz, M. V., Ramírez-Nava, E. J., Hernández-Ochoa, B., Morales-Luna, L., González-Valdez, A., Serrano-Posada, H., Cárdenas-Rodríguez, N., Ortiz-Ramírez, P., Centeno-Leija, S., Arreguin-Espinosa, R., Cuevas-Cruz, M., Ortega-Cuellar, D., Pérez de la Cruz, V., Rocha-Ramírez, L. M., Sierra-Palacios, E., Castillo-Rodríguez, R. A., Baeza-Ramírez, I., Marcial-Quino, J., & Gómez-Manzo, S. (2020). Effects of Single and Double Mutants in Human Glucose-6-Phosphate Dehydrogenase Variants Present in the Mexican Population: Biochemical and Structural Analysis. International Journal of Molecular Sciences, 21(8), 2732. https://doi.org/10.3390/ijms21082732