Phylogenetic Placement of the Plesioclytini (Coleoptera: Cerambycidae: Cerambycinae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling
2.2. Specimens Examined
2.3. Gene Sampling
2.4. Alignment and Tree Reconstruction
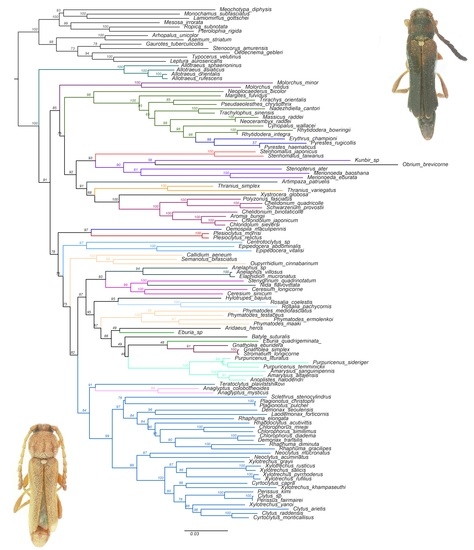
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tavakilian, G.; Chevillotte, H. Base de Donnees Titan sur les Cerambycides ou Longicornes. Available online: http://titan.gbif.fr/aff_sous_famille.php (accessed on 14 November 2021).
- Haddad, S.; Shin, S.; Lemmon, A.R.; Lemmon, E.M.; Svacha, P.; Farrell, B.; Ślipiński, A.; Windsor, D.; Mckenna, D.D. Anchored hybrid enrichment provides new insights into the phylogeny and evolution of longhorned beetles (C erambycidae). Syst. Èntomol. 2018, 43, 68–89. [Google Scholar] [CrossRef]
- Svacha, P.; Lawrence, J.F. 2.4. Cerambycidae Latreille, 1802. In Arthropoda: Insecta: Coleoptera; De Gruyter: Berlin, German, 2014; Volume 3. [Google Scholar]
- Linsley, E.G. The Cerambycidae of North America Part I–III; Linsley, E.G., Steinhaus, E.A., Smith, R.F., Usinger, R.L., Eds.; University of California Press: Berkeley, CA, USA, 1961; Volume 18. [Google Scholar]
- Bouchard, P.; Bousquet, Y.; Davies, A.E.; Alonso-Zarazaga, M.A.; Lawrence, J.F.; Lyal, C.; Newton, A.F.; Reid, C.A.M.; Schmitt, M.; Ślipiński, S.A.; et al. Family-Group Names in Coleoptera (Insecta). ZooKeys 2011, 88, 1–972. [Google Scholar] [CrossRef] [Green Version]
- Leschen, R.A.B.; Colonnelli, E.; Beutel, R.G.; Alonso-Zarazaga, M.A.; Anderson, R.; Bartolozzi, L.; Bezdek, J.; Borowiec, L.; Caldara, R.; Chamorro-Lacayo, L. Morphology and Systematics: Phytophaga. Handbook of Zoology, Arthropoda: Insecta; Coleoptera, Beetles, Volume 3: Morphology and Systematics (Phytophaga); Leschen, R.A.B., Beutel, R.G., Eds.; De Gruyter, Inc.: Berlin, Germany, 2014. [Google Scholar]
- Haddad, S.; McKenna, D.D. Phylogeny and evolution of the superfamily Chrysomeloidea (Coleoptera: Cucujiformia). Syst. Èntomol. 2016, 41, 697–716. [Google Scholar] [CrossRef]
- Napp, D.S. Phylogenetic relationships among the subfamilies of Cerambycidae Coleoptera, Chrysomeloidea). Rev. Bra-Sileira Entomol. 1994, 28, 265–419. [Google Scholar]
- Souza, D.D.S.; Marinoni, L.; Monné, M.L.; Gómez-Zurita, J. Molecular phylogenetic assessment of the tribal classification of Lamiinae (Coleoptera: Cerambycidae). Mol. Phylogenet. Evol. 2020, 145, 106736. [Google Scholar] [CrossRef]
- Wang, J.; Dai, X.-Y.; Xu, X.-D.; Zhang, Z.-Y.; Yu, D.-N.; Storey, K.B.; Zhang, J.-Y. The complete mitochondrial genomes of five longicorn beetles (Coleoptera: Cerambycidae) and phylogenetic relationships within Cerambycidae. PeerJ 2019, 7, e7633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezark, L. Cerambycidae Catalog. A Photographic Catalog of the Cerambycidae of the World. Available online: http://bezbycids.com/byciddb/wdefault.asp?w=n (accessed on 31 October 2021).
- Monne, M.L.; Monne, M.A.; Wang, Q. General Morphology, Classification, and Biology of Cerambycide. In Cerambycidae of the World: Biology and Pest Management; Wang, Q., Ed.; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Yanega, D. Field Guide to Northeastern Longhorned Beetles (Coleoptera: Cerambycidae); Rice, T., Warwick, C., Eds.; Illinois Natural History Survey: Champaign, IL, USA, 1996. [Google Scholar]
- Wappes, J.E.; Skelley, P.E. Description of a new species of Plesioclytus Giesbert (Coleoptera: Cerambycidae: Cerambycinae) from Georgia, and transfer of the genus to Plesioclytini Wappes and Skelley, new tribe. Insecta Mundi 2015, 0436, 1–7. [Google Scholar]
- Linsley, E.G. Taxonomy and classification of the subfamily Cerambycinae, tribes Callichromini through Ancylocerini. In The Cerambycidae of North America Part V; Smith, R.F., Doutt, R.L., Bohart, R.M., Steinhaus, E.A., Usinger, R.L., Eds.; University of California Press: Berkeley, CA, USA, 1964; Volume 22. [Google Scholar]
- Giesbert, E.F. A new genus and species of clytine cerambycid from Florida. Insecta Mundi 1993, 7, 129–131. [Google Scholar]
- Schnepp, K.E. Notes on the natural history of Plesioclytus morrisi Wappes and Skelley, 2015 and Plesioclytus relictus Gies-bert, 1993 (Coleoptera: Cerambycidae). Insecta Mundi 2019, 734, 1–8. [Google Scholar]
- Lee, S.; Lee, S. Multigene phylogeny uncovers oviposition-related evolutionary history of Cerambycinae (Coleoptera: Cerambycidae). Mol. Phylogenet. Evol. 2020, 145, 106707. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Komori, M.; Tsuji-Ueno, S.; Suzuki, M.; Kosaku, A.; Miyamoto, K.; Nishigaki, K. Genome Profiling (GP) Method Based Classification of Insects: Congruence with That of Classical Phenotype-Based One. PLoS ONE 2011, 6, e23963. [Google Scholar] [CrossRef]
- Farrell, B.D.; Blitzer, R.D.; Connor, J.H.; Brown, G.P.; Wong, T.; Shenolikar, S.; Iyengar, R.; Landau, E.M. “Inordinate Fondness” Explained: Why Are There So Many Beetles? Science 1998, 281, 555–559. [Google Scholar] [CrossRef]
- Gomez-Zurita, J.; Hunt, T.; Kopliku, F.; Vogler, A.P. Recalibrated Tree of Leaf Beetles (Chrysomelidae) Indicates Independent Diversification of Angiosperms and Their Insect Herbivores. PLoS ONE 2007, 2, e360. [Google Scholar] [CrossRef] [Green Version]
- Hebert, P.D.N.; Ratnasingham, S.; Zakharov, E.V.; Telfer, A.C.; Levesque-Beaudin, V.; Milton, M.A.; Pedersen, S.; Jannetta, P.; Dewaard, J.R. Counting animal species with DNA barcodes: Canadian insects. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, M.B.; Wingard, S.W.; Szalanski, A.L.; Stephen, F.M. Molecular Diagnostics of Enaphalodes Rufulus (Coleoptera: Cerambycidae). Fla. Èntomol. 2006, 89, 251–256. [Google Scholar] [CrossRef]
- Wu, Y.; Trepanowski, N.F.; Molongoski, J.J.; Reagel, P.F.; Lingafelter, S.W.; Nadel, H.; Myers, S.W.; Ray, A.M. Identification of wood-boring beetles (Cerambycidae and Buprestidae) intercepted in trade-associated solid wood packaging material using DNA barcoding and morphology. Sci. Rep. 2017, 7, 40316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, J.-H.; Roh, S.W.; Whon, T.W.; Jung, M.-J.; Kim, M.-S.; Park, D.-S.; Yoon, C.; Nam, Y.-D.; Kim, Y.-J.; Choi, J.-H.; et al. Insect Gut Bacterial Diversity Determined by Environmental Habitat, Diet, Developmental Stage, and Phylogeny of Host. Appl. Environ. Microbiol. 2014, 80, 5254–5264. [Google Scholar] [CrossRef] [Green Version]
- Grebennikov, V.V.; Jendek, E.; Smirnov, M.E. Diagnostic and phylogenetic utility of the first DNA barcode library for long-horn beetles (Coleoptera: Cerambycidae) from the Russian Far East. Zootaxa 2017, 4276, 441–445. [Google Scholar] [CrossRef]
- Jung, S.W.; Min, H.K.; Kim, Y.-H.; Choi, H.A.; Lee, S.Y.; Bae, Y.J.; Paek, W.K. A DNA barcode library of the beetle reference collection (Insecta: Coleoptera) in the National Science Museum, Korea. J. Asia-Pac. Biodivers. 2016, 9, 234–244. [Google Scholar] [CrossRef] [Green Version]
- Powell, G.S.; Cline, A.R.; Duffy, A.G.; Zaspel, J.M. Phylogeny and reclassification of Carpophilinae (Coleoptera: Nitidulidae), with insights into the origins of anthophily. Zoöl. J. Linn. Soc. 2020, 189, 1359–1369. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sutherland, L.N.; Schnepp, K.E.; Powell, G.S.; Bybee, S.M. Phylogenetic Placement of the Plesioclytini (Coleoptera: Cerambycidae: Cerambycinae). Diversity 2021, 13, 597. https://doi.org/10.3390/d13110597
Sutherland LN, Schnepp KE, Powell GS, Bybee SM. Phylogenetic Placement of the Plesioclytini (Coleoptera: Cerambycidae: Cerambycinae). Diversity. 2021; 13(11):597. https://doi.org/10.3390/d13110597
Chicago/Turabian StyleSutherland, Laura N., Kyle E. Schnepp, Gareth S. Powell, and Seth M. Bybee. 2021. "Phylogenetic Placement of the Plesioclytini (Coleoptera: Cerambycidae: Cerambycinae)" Diversity 13, no. 11: 597. https://doi.org/10.3390/d13110597
APA StyleSutherland, L. N., Schnepp, K. E., Powell, G. S., & Bybee, S. M. (2021). Phylogenetic Placement of the Plesioclytini (Coleoptera: Cerambycidae: Cerambycinae). Diversity, 13(11), 597. https://doi.org/10.3390/d13110597