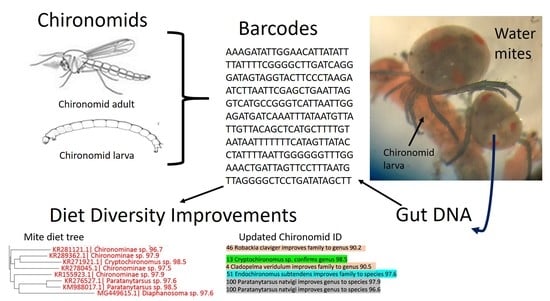
Improved Chironomid Barcode Database Enhances Identification of Water Mite Dietary Content
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling of Chironomids and Water Mites
2.2. Morphological Identification of Chironomids and Water Mites
2.3. DNA Extraction, Amplification, Sequencing
2.4. Bioinformatics of Chironomid Sequences
2.5. Curation of Chironomid Sequences
2.6. Identification of Water Mite Prey Using the Curated Chironomid Database
3. Results
3.1. Chironomid Biodiversity and Barcode Gap Revealed by Morphology and DNA Barcodes
3.2. Pairwise Analysis of Distances between Curated Chironomid Sequences
3.3. Improved Identification of Water Mite Prey Using the Curated Chironomid Sequences Database
4. Discussion
4.1. New Barcodes
4.2. Insights into “Barcode Gaps” in Chironomids
4.3. Improved Water Mite Diet Identifications
Specific Taxa Found in Water Mite Diets
4.4. Need for Further Improvements of Knowledge of Chironomid Diversity
5. Conclusions and Future Considerations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Albert, J.S.; Destouni, G.; Duke-Sylvester, S.M.; Magurran, A.E.; Oberdorff, T.; Reis, R.E.; Winemiller, K.O.; Ripple, W.J. Scientists’ warning to humanity on the freshwater biodiversity crisis. Ambio 2020, 50, 85–94. [Google Scholar] [CrossRef]
- Rapp, T.; Shuman, D.A.; Graeb, B.D.S.; Chipps, S.R.; Peters, E.J. Diet composition and feeding patterns of adult shovelnose sturgeon (Scaphirhynchus platorynchus) in the lower Platte River, Nebraska, USA. J. Appl. Ichthyol. 2011, 27, 351–355. [Google Scholar] [CrossRef]
- Failla, A.; Vasquez, A.; Fujimoto, M.; Ram, J. The ecological, economic and public health impacts of nuisance chironomids and their potential as aquatic invaders. Aquat. Invasions 2015, 10, 1–15. [Google Scholar] [CrossRef]
- Mantilla, J.G.; Gomes, L.; Cristancho, M. The differential expression of Chironomus spp genes as useful tools in the search for pollution biomarkers in freshwater ecosystems. Briefings Funct. Genom. 2017, 17, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Koperski, P. Taxonomic, phylogenetic and functional diversity of leeches (Hirudinea) and their suitability in biological assessment of environmental quality. Knowl. Manag. Aquat. Ecosyst. 2017, 418, 49. [Google Scholar] [CrossRef] [Green Version]
- Hamidoghli, A.; Falahatkar, B.; Khoshkholgh, M.; Sahragard, A. Production and Enrichment of Chironomid Larva with Different Levels of Vitamin C and Effects on Performance of Persian Sturgeon Larvae. North Am. J. Aquac. 2014, 76, 289–295. [Google Scholar] [CrossRef]
- Vasquez, A.A.; Kabalan, B.A.; Ram, J.L.; Miller, C.J. The Biodiversity of Water Mites That Prey on and Parasitize Mosquitoes. Diversity 2020, 12, 226. [Google Scholar] [CrossRef]
- Smith, B.P. Host-parasite interaction and impact of larval water mites on insects. Annu. Rev. Entomol. 1988, 33, 487–507. [Google Scholar] [CrossRef]
- Smith, I.M.; Oliver, D. Review of parasitic associations of larval water mites (acari: Parasitengona: Hydrachnida) with insect hosts. Can. Èntomol. 1986, 118, 407–472. [Google Scholar] [CrossRef]
- Proctor, H.; Pritchard, G. Neglected predators-water mites (Acari, Parasitengona, Hydrachnellae) in fresh-water communities. J. North Am. Benthol. Soc. 1989, 8, 100–111. [Google Scholar] [CrossRef]
- Winkel, E.H.T.; Davids, C.; De Nobel, J. Food and Feeding Strategies of Water Mites of the Genus Hygrobates and the Impact of Their Predation on the Larval Population of the Chironomid Cladotanytarsus Mancus (Walker) in Lake Maarsseveen. Neth. J. Zool. 1988, 39, 246–263. [Google Scholar] [CrossRef]
- Shatrov, A.B. Anatomy and ultrastructure of the salivary (prosomal) glands in unfed water mite larvae Piona carnea (C.L. Koch, 1836) (Acariformes: Pionidae). Zool. Anz.-A J. Comp. Zool. 2012, 251, 279–287. [Google Scholar] [CrossRef]
- Cohen, A.C. Solid-to-Liquid Feeding: The Inside(s) Story of Extra-Oral Digestion in Predaceous Arthropoda. Am. Èntomol. 1998, 44, 103–117. [Google Scholar] [CrossRef]
- Martin, P.; Koester, M.; Schynawa, L.; Gergs, R. First detection of prey DNA in Hygrobates fluviatilis (Hydrachnidia, Acari): A new approach for determining predator-prey relationships in water mites. Exp. Appl. Acarol. 2015, 67, 373–380. [Google Scholar] [CrossRef]
- Vasquez, A.A.; Mohiddin, O.; Li, Z.; Bonnici, B.L.; Gurdziel, K.; Ram, J.L. Molecular diet studies of water mites reveal prey biodiversity. PLoS ONE 2021, 16, e0254598. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Failla, A.; Vasquez, A.; Hudson, P.; Fujimoto, M.; Ram, J. Morphological identification and COI barcodes of adult flies help determine species identities of chironomid larvae (Diptera, Chironomidae). Bull. Èntomol. Res. 2015, 106, 34–46. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Vasquez, A.A.; Hudson, P.L.; Fujimoto, M.; Keeler, K.; Armenio, P.M.; Ram, J.L. Eurytemora carolleeae in the Laurentian Great Lakes revealed by phylogenetic and morphological analysis. J. Great Lakes Res. 2016, 42, 802–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasquez, A.A.; Qazazi, M.S.; Fisher, J.R.; Failla, A.J.; Rama, S.; Ram, J.L. New molecular barcodes of water mites (Trombidiformes: Hydrachnidiae) from the Toledo Harbor region of Western Lake Erie, USA, with first barcodes forKrendowskia(Krendowskiidae) andKoenikea(Unionicolidae). Int. J. Acarol. 2017, 43, 494–498. [Google Scholar] [CrossRef]
- Vasquez, A.A.; Carmona-Galindo, V.; Qazazi, M.S.; Walker, X.N.; Ram, J.L. Water mite assemblages reveal diverse genera, novel DNA barcodes and transitional periods of intermediate disturbance. Exp. Appl. Acarol. 2020, 80, 491–507. [Google Scholar] [CrossRef]
- Townes, H.K. The nearctic species of Tendipedini-Diptera, Tendipedidae (=chironomidae). Am. Midl. Nat. 1945, 34, 1–206. [Google Scholar] [CrossRef]
- Saether, O.A. Glyptotendipes Kieffer and Demeijerea Kruseman from Lake Winnipeg, Manitoba, Canada, with the description of four new species (Diptera: Chironomidae). Zootaxa 2011, 2760, 39–52. [Google Scholar] [CrossRef] [Green Version]
- Saether, O.A. Cryptochironomus Kieffer from Lake Winnipeg, Canada, with a review of Nearctic species (Diptera: Chironomidae). Zootaxa 2009, 2208, 1–24. [Google Scholar] [CrossRef]
- Epler, J.H. Biosystematics of the genus Dicrotendipes Kieffer, 1913 (Diptera: Chironomidae) of the world. Mem. Am. Entomol. Soc. 1988, 36, 1–214. [Google Scholar]
- Dendy, J.S.; Sublette, J.E. The Chironomidae (=Tendipedidae: Diptera) of Alabama with Descriptions of Six New Species1. Ann. Èntomol. Soc. Am. 1959, 52, 506–519. [Google Scholar] [CrossRef]
- Cranston, P.S.; Dillon, M.E.; Pinder, L.C.V.; Reiss, F. The adult males of Chironominae (Diptera, Chironomidae) of the holarctic region-keys and diagnoses. Entomol. Scand. 1989, 34, 353–502. [Google Scholar]
- Roback, S.S. Monograph 17 the academy of natural sciences of Philadelphia the adults of the subfamily Tanypodinae equals pelopiinae in North America Diptera chironomidae. Monogr. Acad. Nat. Sci. Phila. 1971, 17, 410. [Google Scholar]
- Heyn, M.W. A review of the systematic position of the North American species of the genusGlyptotendipes. Aquat. Ecol. 1992, 26, 129–137. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Epler, J.H. Revision of the Nearctic Dicrotendipes Kieffer, 1913 (Diptera: Chironomidae). Evol. Monogr. 1987, 9, 1–102. [Google Scholar]
- Saether, O.A. Taxonomic studies on Chironomidae Nanocladius pseudochironomus and the Harnischia complex. Bull. Fish. Res. Board Can. 1977, 196, 1–141. [Google Scholar]
- Hudson, P.L.; Lenat, D.R.; Caldwell, B.A.; Smith, D. Chironomidae of the Southeastern United States: A Checklist of Species and Notes on Biology, Distribution, and Habitat; U S Fish and Wildlife Service Fish and Wildlife Research: Washington, DC, USA, 1990; pp. 1–46. [Google Scholar]
- Winnell, M.H.; Jude, D.J. Associations among Chironomidae and Sandy Substrates in Nearshore Lake Michigan. Can. J. Fish. Aquat. Sci. 1984, 41, 174–179. [Google Scholar] [CrossRef]
- Yan, C.; Wang, X. Robackia Saether from China (Diptera: Chironomidae). Zootaxa 2006, 1361, 53–59. [Google Scholar] [CrossRef]
- Hudson, P.L. Unusual larval habitats and life-history of Chironomid (Diptera) genera. Entomol. Scand. 1987, 29, 369–373. [Google Scholar]
- Roback, S. Adults of the Subfamily Tanypodinae (-Pelopinae) in North America (Diptera: Chironomidae); Academy of Natural Sciences: Hinckley, MN, USA, 2007; Volume 17, p. 410. [Google Scholar]
- Aydın, G.B. The growth of Tanypus punctipennis meigen (Diptera, Chironomidae) larvae in laboratory conditions and the effects of water temperature and ph. Trak. Univ. J. Nat. Sci. 2018, 19, 101–105. [Google Scholar] [CrossRef]
- Specziár, A. Life history patterns of Procladius choreus, Tanypus punctipennis and Chironomus balatonicus in Lake Balaton. Ann. de Limnol.-Int. J. Limnol. 2008, 44, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Stur, E.; da Silva, F.L.; Ekrem, T. Back from the Past: DNA Barcodes and Morphology Support Ablabesmyia americana Fittkau as a Valid Species (Diptera: Chironomidae). Diversity 2019, 11, 173. [Google Scholar] [CrossRef] [Green Version]
- Int Panis, L.; Boudewijn, G.; Lieven, B.; Verheyen, R.F. Ablabesmyia longistyla Fittkau, 1962 (Diptera: Chironomidae), new for the Belgian fauna. Bull. Annls Soc. R. Belg. Ent. 1992, 128, 316–318. [Google Scholar]
- Oliver, D.R.; Roussel, M.E. The Genera of Larval Midges of Canada Diptera: Chironomidae; Insectes et Arachnides du Canada; Research Branch, Agriculture Canada: Ottawa, ON, Canada, 1983; pp. 1–263. [Google Scholar]
- Egan, A.T.; Ferrington, L.C., Jr. Chironomidae (Diptera) in Freshwater Coastal Rock Pools at Isle Royale, Michigan. Trans. Am. Èntomol. Soc. 2015, 141, 1–25. [Google Scholar] [CrossRef]
- Hudson, P.L.; Adams, J.V. Sieve efficiency in benthic sampling as related to chironomid head capsule width. J. Kans. Entomol. Soc. 1998, 71, 456–468. [Google Scholar]
- Roback, S.S.; Bereza, D.J.; Vidrine, M.F. Description of an Ablabesmyia [Diptera: Chironomidae: Tanypodinae] Symbiont of Unionid Fresh-Water Mussels [Mollusca:Bivalvia:Unionacea], with Notes on Its Biology and Zoogeography. Trans. Am. Entomol. Soc. 1979, 105, 577–620. [Google Scholar]
- Kaster, J.L.; Bushnell, J.H. Occurrence of Tests and Their Possible Significance in the Worm, Tubifex tubifex (Oligochaeta). Southwest. Nat. 1981, 26, 307. [Google Scholar] [CrossRef]
- Oliveira, C.S.N.; Fonseca-Gessner, A.A.; Silva, M.A.N. The immature stages of Ablabesmyia (Sartaia) metica Roback, 1983 (Diptera: Chironomidae) with keys to subgenera. Zootaxa 2008, 1808, 61–68. [Google Scholar] [CrossRef]
- Roback, S.S. Ablabesmyia (Sartaia) metica, a New Subgenus and Species (Diptera: Chironomidae: Tanypodinae). Proc. Acad. Nat. Sci. Phila. 1983, 135, 236–240. [Google Scholar]
- Beck, W.M. Biology of the larval chironomids. State Fla. Dep. Environ. Regul. 1976, 2, 58. [Google Scholar]
- Boesel, M.W. The early stages of Ablabesmyia annulata (Say) (Diptera, Chironomidae). Ohio J. Sci. 1972, 72, 3. [Google Scholar]
- Pfenninger, M.; Nowak, C.; Kley, C.; Steinke, D.; Streit, B. Utility of DNA taxonomy and barcoding for the inference of larval community structure in morphologically cryptic Chironomus (Diptera) species. Mol. Ecol. 2007, 16, 1957–1968. [Google Scholar] [CrossRef] [PubMed]
- Brodin, Y.; Ejdung, G.; Strandberg, J.; Lyrholm, T. Improving environmental and biodiversity monitoring in the Baltic Sea using DNA barcoding of Chironomidae (Diptera). Mol. Ecol. Resour. 2012, 13, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.; Brodnicke, O.; Evankow, A.; Ferreira, A.; Fontes, J.; Hansen, A.; Jensen, M.; Kalaycı, T.; Leeper, A.; Patil, S.; et al. The Future of DNA Barcoding: Reflections from Early Career Researchers. Diversity 2021, 13, 313. [Google Scholar] [CrossRef]
- Elías-Gutiérrez, M.; Jerónimo, F.M.; Ivanova, N.V.; Valdez-Moreno, M.; Hebert, P.D.N. DNA barcodes for Cladocera and Copepoda from Mexico and Guatemala, highlights and new discoveries. Zootaxa 2008, 1839, 1–42. [Google Scholar] [CrossRef] [Green Version]








| Previous GenBank Identification in Mite Diet | Improved Identification |
|---|---|
| Chironomidae | Dicrotendipes modestus |
| Chironomidae | Parachironomus abortivus |
| Chironominae | Endochironomus subtendens |
| Chironomidae | Cricotopus festivellus |
| Chironomidae | Tanypus punctipennis |
| Chironomidae | Paratanytarsus nr. bituberculatus |
| Previous GenBank Identification in Mite Diet | Improved Identification |
|---|---|
| Parachironomus sp. | Parachironomus tenuicaudatus |
| Cricotopus sp. 1 | Cricotopus sylvestris |
| Cricotopus sp. 1 | Cricotopus sylvestris |
| Orthocladius sp. | Orthocladius obumbratus |
| Rheotanytarsus sp. | Rheotanytarsus exiguus gr. |
| Previous GenBank. Identification in Mite Diet | Improved Identification 1 |
|---|---|
| Chironominae | Cladopelma veridulum |
| Chironomidae | Polypedilum simulans |
| Chironominae | Robackia claviger |
| Chironomidae | Ablabesmyia mallochi |
| Chironomidae | Ablabesmyia annulata |
| Chironomidae | Tanytarsus glabrescens |
| Chironominae | Chironomus sp. |
| Chironomidae | Cricotopus bicinctus gr. |
| Chironomidae | Polypedilum cf. halterale |
| Chironomidae | Polypedilum halterale gr. |
| Chironominae | Polypedilum trigonum |
| Chironomidae | Polypedilum scalaenum |
| Chironomids | OTU Number | Water Mite Species | ||||
|---|---|---|---|---|---|---|
| Chironomid Name | Lebertia davidcooki | Lebertia quinquemaculosa | Lebertia sp. | Limnesia sp. | Arrenurus sp. | |
| Glyptotendipes meridionalis * | 3 | ✓ | ||||
| Cladopelma veridulum * | 4 | ✓ | ✓ | ✓ | ✓ | ✓ |
| Parachironomus sp. * | 6 | ✓ | ✓ | ✓ | ||
| Parachironomus tenuicaudatus | 7 | ✓ | ||||
| Parachironomus hazelriggi * | 8 | ✓ | ||||
| Cryptochironomus sp. * | 13 | ✓ | ✓ | ✓ | ||
| Cryptochironomus ponderosus * | 14 | ✓ | ✓ | |||
| Glypotendipes senilis | 21 | ✓ | ||||
| Dicrotendipes modestus | 22 | ✓ | ✓ | |||
| Chironomus riparius | 28 | ✓ | ✓ | ✓ | ✓ | |
| Chironomus riparius * | 29 | ✓ | ||||
| Parachironomus abortivus | 33 | ✓ | ||||
| Chironomus entis/plumosus | 36 | ✓ | ✓ | |||
| Chironomus maturus | 38 | ✓ | ✓ | |||
| Chironomus sp. * | 39 | ✓ | ||||
| Chironomus crassicaudatus | 42 | ✓ | ✓ | ✓ | ||
| Paratanytarsus sp. * | 43 | ✓ | ✓ | ✓ | ✓ | |
| Polypedilum simulans * | 44 | ✓ | ✓ | |||
| Robackia claviger * | 46 | ✓ | ✓ | |||
| Polypedilum illinoense * | 47 | ✓ | ||||
| Polypedilum scaleneum * | 49 | ✓ | ||||
| Endochironomus subtendens | 51 | ✓ | ✓ | ✓ | ||
| Polypedilum halterale* | 54 | ✓ | ||||
| Polypedilum halterale gr. * | 55 | ✓ | ||||
| Polypedilum trigonum * | 57 | ✓ | ✓ | |||
| Cricotopus sylvestris | 58 | ✓ | ✓ | ✓ | ✓ | |
| Cricotopus sylvestris | 59 | ✓ | ✓ | ✓ | ✓ | |
| Cricotopus festivellus | 60 | ✓ | ✓ | ✓ | ✓ | |
| Cricotopus bicinctus gr. * | 63 | ✓ | ✓ | |||
| Ablabesmyia mallochi * | 70 | ✓ | ||||
| Ablabesmyia annulate * | 72 | ✓ | ||||
| Orthocladius obumbratus | 74 | ✓ | ✓ | |||
| Tanypus punctipennis | 76 | ✓ | ||||
| Coelotanypus sp. * | 82 | ✓ | ||||
| Tanytarsus glabrescens * | 87 | ✓ | ✓ | |||
| Rheotanytarsus exiguous gr. | 91 | ✓ | ||||
| Paratanytarsus nr. bituberculatus | 96 | ✓ | ✓ | ✓ | ||
| Paratanytarsus natvigi * | 100 | ✓ | ✓ | ✓ | ✓ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasquez, A.A.; Bonnici, B.L.; Yusuf, S.H.; Cruz, J.I.; Hudson, P.L.; Ram, J.L. Improved Chironomid Barcode Database Enhances Identification of Water Mite Dietary Content. Diversity 2022, 14, 65. https://doi.org/10.3390/d14020065
Vasquez AA, Bonnici BL, Yusuf SH, Cruz JI, Hudson PL, Ram JL. Improved Chironomid Barcode Database Enhances Identification of Water Mite Dietary Content. Diversity. 2022; 14(2):65. https://doi.org/10.3390/d14020065
Chicago/Turabian StyleVasquez, Adrian A., Brittany L. Bonnici, Safia Haniya Yusuf, Janiel I. Cruz, Patrick L. Hudson, and Jeffrey L. Ram. 2022. "Improved Chironomid Barcode Database Enhances Identification of Water Mite Dietary Content" Diversity 14, no. 2: 65. https://doi.org/10.3390/d14020065
APA StyleVasquez, A. A., Bonnici, B. L., Yusuf, S. H., Cruz, J. I., Hudson, P. L., & Ram, J. L. (2022). Improved Chironomid Barcode Database Enhances Identification of Water Mite Dietary Content. Diversity, 14(2), 65. https://doi.org/10.3390/d14020065