Talarolides Revisited: Cyclic Heptapeptides from an Australian Marine Tunicate-Associated Fungus, Talaromyces sp. CMB-TU011
Abstract
:1. Introduction
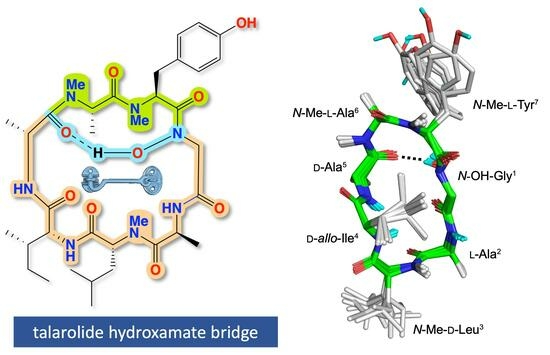
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Collection and Taxonomy of Talaromyces sp. CMB-TU011
3.3. Cultivation and Fractionation of Talaromyces sp. CMB-TU011
3.4. Marfey’s Analysis of Talarolides A–D
3.4.1. Standard Marfey’s Hydrolysis and Derivatization Method #1
3.4.2. Standard Marfey’s HPLC Method #2
3.4.3. Marfey’s HPLC Method #3 Optimized for Resolving N-Me-Ala Derivatives
3.4.4. Marfey’s HPLC Method #4 Optimized for Resolving Leu, Ile and allo-Ile Derivatives
3.4.5. Marfey’s Analysis of Talarolides A–D (1–4)
3.5. Two-Dimensional Marfey’s Analysis of Talarolide B
3.5.1. Two-Dimensional Marfey’s Method #5 Partial Hydrolysis of Talarolide B
3.5.2. Synthesis of Dipeptides l-FDAA-d-allo-Ile-l-Ala and l-FDAA-d-allo-Ile-d-Ala
3.5.3. Coupling of the First Amino Acid
3.5.4. Coupling of Fmoc-d-allo-Ile
3.5.5. Derivatization with l-FDAA
3.5.6. Cleavage of l-FDAA Derivatized Dipeptide from Resin
3.5.7. Marfey’s Method #6 Optimized for l-FDAA-d-allo-Ile-Ala Diastereomers
3.6. Synthesis of Talarolide B
3.6.1. Coupling of the First Amino Acid
3.6.2. Elongation of Peptide Sequence
3.6.3. Cleavage of Linear Protected Peptide
3.6.4. Cyclization of Linear Protected Peptide
3.6.5. Cyclic Peptide Deprotection
3.7. Three-Dimensional Solution Structure Calculations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raju, R.; Khalil, Z.G.; Piggott, A.M.; Blumenthal, A.; Gardiner, D.L.; Skinner-Adams, T.S.; Capon, R.J. Mollemycin A: An Antimalarial and Antibacterial Glyco-Hexadepsipeptide-Polyketide from an Australian Marine-Derived Streptomyces sp. (CMB-M0244). Org. Lett. 2014, 16, 1716–1719. [Google Scholar] [CrossRef] [PubMed]
- Khalil, Z.G.; Salim, A.A.; Lacey, E.; Blumenthal, A.; Capon, R.J. Wollamides: Antimycobacterial Cyclic Hexapeptides from an Australian Soil Streptomyces. Org. Lett. 2014, 16, 5120–5123. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.-H.; Khalil, Z.; Dewapriya, P.; Salim, A.A.; Lin, H.-W.; Capon, R.J. Trichodermides A-E: New Peptaibols Isolated from the Australian Termite Nest-Derived Fungus Trichoderma virens CMB-TN16. J. Nat. Prod. 2018, 81, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Quezada, M.; Shang, Z.; Kalansuriya, P.; Salim, A.A.; Lacey, E.; Capon, R.J. Waspergillamide A, a Nitro Depsi-Tetrapeptide Diketopiperazine from an Australian Mud Dauber Wasp-Associated Aspergillus sp. (CMB-W031). J. Nat. Prod. 2017, 80, 1192–1195. [Google Scholar] [CrossRef] [PubMed]
- Elbanna, A.H.; Khalil, Z.G.; Bernhardt, P.V.; Capon, R.J. Scopularides Revisited: Molecular Networking Guided Exploration of Lipodepsipeptides in Australian Marine Fish Gastrointestinal Tract-Derived Fungi. Mar. Drugs 2019, 17, 475. [Google Scholar] [CrossRef] [PubMed]
- Dewapriya, P.; Khalil, Z.G.; Prasad, P.; Salim, A.A.; Cruz-Morales, P.; Marcellin, E.; Capon, R.J. Talaropeptides A-D: Structure and Biosynthesis of Extensively N-Methylated Linear Peptides from an Australian Marine Tunicate-Derived Talaromyces sp. Front. Chem. 2018, 6, 394. [Google Scholar] [CrossRef] [PubMed]
- Dewapriya, P.; Prasad, P.; Damodar, R.; Salim, A.A.; Capon, R.J. Talarolide A, a Cyclic Heptapeptide Hydroxamate from an Australian Marine Tunicate-Associated Fungus, Talaromyces sp. (CMB-TU011). Org. Lett. 2017, 19, 2046–2049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; De Leon Rodriguez, L.M.; Huang, R.; Leung, I.K.H.; Harris, P.W.R.; Brimble, M.A. Total Synthesis of the Proposed Structure of Talarolide, A. Org. Biomol. Chem. 2018, 16, 5286–5293. [Google Scholar] [CrossRef] [PubMed]
- Salim, A.A.; Khalil, Z.G.; Elbanna, A.H.; Wu, T.; Capon, R.J. Methods in Microbial Biodiscovery. Mar. Drugs 2021, 19, 503. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible Molecular Networking of Untargeted Mass Spectrometry Data Using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Vijayasarathy, S.; Prasad, P.; Fremlin, L.J.; Ratnayake, R.; Salim, A.A.; Khalil, Z.; Capon, R.J. C3 and 2D C3 Marfey’s Methods for Amino Acid Analysis in Natural Products. J. Nat. Prod. 2016, 79, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. Charmm: A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar] [CrossRef]
- Brunger, A.T. X-Plor Manual Version 3.1; Yale University Press: New Haven, CT, USA, 1992. [Google Scholar]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The Chemical Shift Index: A Fast and Simple Method for the Assignment of Protein Secondary Structure through NMR Spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Pardi, A.; Billeter, M.; Wüthrich, K. Calibration of the Angular Dependence of the Amide Proton-Cα Proton Coupling Constants, 3JHNα, in a Globular Protein: Use of 3JHNα for Identification of Helical Secondary Structure. J. Mol. Biol. 1984, 180, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.S.; Hoang, H.N.; Lohman, R.-J.; Hill, T.A.; Lucke, A.J.; Craik, D.J.; Edmonds, D.J.; Griffith, D.A.; Rotter, C.J.; Ruggeri, R.B.; et al. Improving on Nature: Making a Cyclic Heptapeptide Orally Bioavailable. Angew. Chem. 2014, 53, 12059–12063. [Google Scholar] [CrossRef] [PubMed]









| 1 δH, Mult, (J in Hz) | 2 δH, Mult, (J in Hz) e | 3 δH, Mult, (J in Hz) | 4 δH, Mult, (J in Hz) | |
|---|---|---|---|---|
| N-OH-Gly1/Gly1 | ||||
| 2a | 4.75, d (17.1) | 4.14 a | 4.75 a | 4.80 d (17.0) |
| 2b | 3.76, d (17.1) | 3.54, dd (17.3, 2.9) | 3.76, d (17.2) | 3.71, d (17.0) |
| N-OH | 9.31, s | 9.41, s | 9.13, s | |
| N-H | 7.53, dd (7.6, 2.9) | |||
| l-Ala2 | ||||
| 2 | 4.49, qd (6.8, 4.1) | 4.67, qd (6.7, 5.9) | 4.50, qd (6.7, 4.0) | 4.49, qd (6.8, 4.1) |
| 3 | 1.19, d (6.8) | 1.20, d (6.7) | 1.20, d (6.7) | 1.19, d (6.8) |
| N-H | 8.65, d (4.1) | 8.59, d (5.9) | 8.65, d (4.0) | 8.62, d (4.1) |
| N-Me-d-Leu3 | ||||
| 2 | 5.05, dd (11.8, 3.9) | 5.05, dd (11.8, 3.8) | 5.07, dd (11.6, 3.8) | 5.06, dd (11.7, 3.8) |
| 3a | 1.79, ddd (14.4, 10.3, 3.9) | 1.82, ddd (14.4, 10.5, 3.8) | 1.79, ddd (14.4, 10.3, 3.8) | 1.79, ddd (13.4, 10.6, 3.9) |
| 3b | 1.58, ddd (14.4, 11.8, 3.9) | 1.61, ddd (14.4, 11.8, 3.7) | 1.57, ddd (14.4, 11.6, 3.9) | 1.57, ddd (13.4, 11.7, 3.9) |
| 4 | 1.37, m | 1.39, m | 1.38, m | 1.38 a |
| 5 | 0.77, d (6.5) | 0.79, d (6.5) | 0.78, d (6.5) | 0.77, d (6.5) |
| 6 | 0.88, d (6.5) | 0.90, d (6.5) | 0.89, d (6.5) | 0.88, d (6.6) |
| N-Me | 3.00, s | 3.11, s | 3.01, s | 2.98, s |
| d-allo-Ile4/d-Val4 | ||||
| 2 | 4.72 a | 4.60, dd (9.3, 5.2) | 4.58, dd (9.5, 4.9) | 4.69, dd (9.5, 3.9) |
| 3 | 1.95, m | 1.95, m | 2.17, m | 1.91, m |
| 4a | 1.42, m | 1.56, m | 0.92, d (6.8) | 1.39 a |
| 4b | 1.07, m | 1.09 b | 1.05, m | |
| 5 | 0.94, dd (7.3, 7.3) | 0.95, dd (7.3, 7.3) | 0.86, d (6.8) | 0.92, dd (7.3, 7.3) |
| 6 | 0.81, d (6.9) | 0.92, d (6.9) | 0.80, d (6.9) | |
| N-H | 7.24, d (9.6) | 6.96, d (9.3) | 7.23, d (9.5) | 7.14, d (9.6) |
| d-Ala5 | ||||
| 2 | 4.34, qd (7.1, 5.4) | 4.42 c | 4.33, qd (7.1, 5.0) | 3.90, m |
| 3 | 1.12, d (7.1) | 1.10 b, d (7.1) | 1.12, d (7.1) | 1.13, d (7.1) |
| N-H | 8.87, d (5.4) | 8.63, d (5.4) | 8.87, d (5.0) | 8.58, d (5.2) |
| N-Me-l-Ala/l-Ala6 | ||||
| 2 | 4.71 a | 4.43 c | 4.75 a | 4.37, m |
| 3 | 0.49, d (6.5) | 0.58, d (6.5) | 0.52, d (6.5) | 0.63, d (6.4) |
| N-Me | 2.70, s | 2.70, s | 2.71, s | |
| N-H | 8.27, d (9.5) | |||
| N-Me-l-Tyr7 | ||||
| 2 | 4.80, dd (10.5, 4.9) | 4.15 a | 4.80, dd (9.8, 5.3) | 5.01, dd (8.6, 6.6) |
| 3a | 2.84, dd (14.3, 10.5) | 2.93, dd (14.1, 11.6) | 2.82, dd (14.2, 9.8) | 2.81, dd (14.3, 6.6) |
| 3b | 2.60, dd (14.3, 4.9) | 2.55 d | 2.62, dd (14.2, 5.3) | 2.77, dd (14.3, 8.6) |
| 5/9 | 6.93, d (8.4) | 6.90, d (8.4) | 6.94, d (8.4) | 6.99, d (8.4) |
| 6/8 | 6.64, d (8.4) | 6.63, d (8.4) | 6.63, d (8.4) | 6.63, d (8.4) |
| 7-OH | 9.20, s | 9.25, s | 9.18, s | 9.15, s |
| N-Me | 2.66, s | 2.86, s | 2.67, s | 2.64, s |
| 1 δC | 2 δC | 3 δC | 4 δC | |
|---|---|---|---|---|
| N-OH-Gly1/Gly1 | ||||
| 1 | 167.3 | 169.3 | 167.3 | 167.5 |
| 2 | 50.2 | 41.3 | 50.1 | 49.9 |
| l-Ala2 | ||||
| 1 | 174.1 | 174.4 | 174.1 | 174.0 |
| 2 | 45.2 | 44.6 | 45.2 | 45.2 |
| 3 | 15.7 | 15.8 | 15.7 | 15.7 |
| N-Me-d-Leu3 | ||||
| 1 | 169.5 | 169.2 a | 169.5 | 169.5 |
| 2 | 54.5 | 54.6 b | 54.4 | 54.4 |
| 3 | 36.0 | 36.1 | 35.9 | 36.0 |
| 4 | 24.4 | 24.4 | 24.4 | 24.4 |
| 5 | 21.0 | 20.9 | 20.9 | 20.9 |
| 6 | 23.3 | 23.3 | 23.3 | 23.3 |
| N-Me | 31.0 | 30.8 | 31.0 | 30.9 |
| d-allo-Ile4/d-Val4 | ||||
| 1 | 172.0 | 171.2 | 171.6 | 171.8 |
| 2 | 53.7 | 54.6 b | 55.3 | 53.8 |
| 3 | 38.5 | 38.5 | 31.9 | 38.8 |
| 4 | 26.2 | 25.7 | 19.4 | 26.4 |
| 5 | 12.0 | 12.0 | 17.0 | 11.9 |
| 6 | 13.7 | 14.2 c | 13.7 | |
| d-Ala5 | ||||
| 1 | 171.1 | 171.6 | 171.0 | 170.8 |
| 2 | 45.8 | 45.7 | 45.8 | 48.6 |
| 3 | 14.9 | 14.6 | 14.9 | 16.7 |
| N-Me-l-Ala/l-Ala6 | ||||
| 1 | 169.8 | 169.8 | 169.8 | 171.4 |
| 2 | 46.7 | 49.3 | 46.7 | 42.7 |
| 3 | 15.1 | 14.2 c | 15.1 | 18.5 |
| N-Me | 28.6 a | 29.1 | 28.6 a | |
| N-Me-l-Tyr7 | ||||
| 1 | 168.2 | 169.2 a | 168.2 | 168.3 |
| 2 | 56.6 | 59.8 | 56.6 | 57.1 |
| 3 | 34.1 | 34.8 | 34.3 | 34.5 |
| 4 | 126.6 | 126.5 | 126.7 | 127.2 |
| 5/9 | 130.8 | 130.3 | 130.7 | 130.6 |
| 6/8 | 114.8 | 114.9 | 114.8 | 114.9 |
| 7 | 155.9 | 156.0 | 155.9 | 155.8 |
| N-Me | 28.6 a | 29.5 | 28.6 a | 28.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salim, A.A.; Hussein, W.M.; Dewapriya, P.; Hoang, H.N.; Zhou, Y.; Samarasekera, K.; Khalil, Z.G.; Fairlie, D.P.; Capon, R.J. Talarolides Revisited: Cyclic Heptapeptides from an Australian Marine Tunicate-Associated Fungus, Talaromyces sp. CMB-TU011. Mar. Drugs 2023, 21, 487. https://doi.org/10.3390/md21090487
Salim AA, Hussein WM, Dewapriya P, Hoang HN, Zhou Y, Samarasekera K, Khalil ZG, Fairlie DP, Capon RJ. Talarolides Revisited: Cyclic Heptapeptides from an Australian Marine Tunicate-Associated Fungus, Talaromyces sp. CMB-TU011. Marine Drugs. 2023; 21(9):487. https://doi.org/10.3390/md21090487
Chicago/Turabian StyleSalim, Angela A., Waleed M. Hussein, Pradeep Dewapriya, Huy N. Hoang, Yahao Zhou, Kaumadi Samarasekera, Zeinab G. Khalil, David P. Fairlie, and Robert J. Capon. 2023. "Talarolides Revisited: Cyclic Heptapeptides from an Australian Marine Tunicate-Associated Fungus, Talaromyces sp. CMB-TU011" Marine Drugs 21, no. 9: 487. https://doi.org/10.3390/md21090487
APA StyleSalim, A. A., Hussein, W. M., Dewapriya, P., Hoang, H. N., Zhou, Y., Samarasekera, K., Khalil, Z. G., Fairlie, D. P., & Capon, R. J. (2023). Talarolides Revisited: Cyclic Heptapeptides from an Australian Marine Tunicate-Associated Fungus, Talaromyces sp. CMB-TU011. Marine Drugs, 21(9), 487. https://doi.org/10.3390/md21090487