Coumarin Derivatives as New Toxic Compounds to Selected K12, R1–R4 E. coli Strains
Abstract
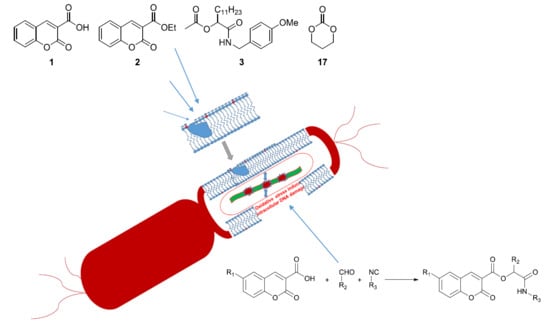
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Antimicrobial Properties of the Used Compounds
2.3. Modification of Plasmid DNA Isolated from E. coli R1–R4 Strains with Tested Coumarin Derivatives
3. Conclusions
4. Experimental Section
4.1. Strains and Media
4.2. Experimental Chemistry
4.3. General Procedure for Synthesis of Compounds 3–17
4.3.1. 1-(4-Methoxybenzylamino)-1-Oxotridecan-2-yl Acetate 3
4.3.2. 1-(4-Methoxybenzylamino)-1-Oxotridecan-2-yl-3-Coumarincarboxylate 4
4.3.3. 1-(4-Methoxybenzylamino)-1-Oxotridecan-2-yl-3-(6-Nitrocoumarin) Carboxylate 5
4.3.4. 1-(4-Methoxybenzylamino)-1-Oxotridecan-2-yl-3-(6 Methoxycoumarin) Carboxylate, 6
4.3.5. 1-(4-Methoxybenzylamino)-1-Oxotridecan-2-yl-3-(6-Methylcoumarin) Carboxylate 7
4.3.6. 1-(Benzylamino)-1-Oxotridecan-2-yl-3-Coumarincarboxylate 8
4.3.7. 1-(t-Butylamino)-1-Oxotridecan-2-yl-3-Coumarincarboxylate 9
4.3.8. 1-(Cyclohexylamino)-1-Oxotridecan-2-yl-3-Coumarincarboxylate 10
4.3.9. Ethyl 2-[2-(3-Coumarincarboxylate)-1-Oxotridecanamino]acetate 11
4.3.10. 1-(4-Methoxybenzylamino)-1-Oxononan-2-yl-3-Coumarincarboxylate 12
4.3.11. 1-(4-Methoxybenzylamino)-1-Oxoheptan-2-yl-3-Coumarincarboxylate 13
4.3.12. 1-(4-Methoxybenzylamino)-1-Oxobutan-2-yl-3-Coumarincarboxylate 14
4.3.13. 1-(4-Methoxybenzylamino)-1-oxo-3-Phenylpropan-2-yl-3-Coumarincarboxylate 15
4.3.14. 1-(4-Methoxybenzylamino)-3-Methyl-1-Oxobutan-2-yl-3-Coumarincarboxylate 16
4.4. Estimation of Minimum Inhibition Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
4.5. Interaction of the Plasmid DNA with Peptidomimetics
4.6. Statistical Analysis.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MIC | Minimum inhibitory concentration |
| MBC | Minimum bactericidal concentration |
| oc | open circle |
| ccc | covalently-closed circle |
References
- Marquarding, D.; Gokel, G.; Hoffmann, P.; Ugi, I. Chapter 7—The Passerini Reaction and Related Reactions. Org. Chem. 1971, 20, 133. [Google Scholar] [CrossRef]
- Bos, M.; Riguet, E. Synthesis of Chiral γ-Lactones by One-Pot Sequential Enantioselective Organocatalytic Michael Addition of Boronic Acids and Diastereoselective Intramolecular Passerini Reaction. J. Org. Chem. 2014, 79, 10881–10889. [Google Scholar] [CrossRef] [PubMed]
- Bossio, R.; Marcaccini, S.; Pepino, R.; Torroba, T. Synthesis Studies on Isocyanides and Related Compounds: A Novel Synthetic Route to Furan Derivatives. Heterocycles 1993, 783. [Google Scholar] [CrossRef]
- Szymanski, W.; Ostaszewski, R. Toward stereocontrolled, chemoenzymatic synthesis of unnatural peptides. Tetrahedron 2008, 64, 3197–3203. [Google Scholar] [CrossRef]
- Szymanski, W.; Zwolińska, M.; Ostaszewski, R. Studies on the application of the Passerini reaction and enzymatic procedures to the synthesis of tripeptide mimetics. Tetrahedron 2007, 63, 7647–7653. [Google Scholar] [CrossRef]
- Szymanski, W.; Ostaszewski, R. Multicomponent diversity and enzymatic enantioselectivity as a route towards both enantiomers of α-amino acids—A model study. Tetrahedron Asymmetry 2006, 17, 2667–2671. [Google Scholar] [CrossRef]
- Dömling, A.; Ugi, I. IMulticomponent Reactions with Isocyanides. Angew. Chem. Int. Ed. Engl. 2000, 39, 3168–3210. [Google Scholar] [CrossRef]
- Das, K.; Jain, B.; Gupta, P. Photophysics of Coumarin 500 and Coumarin 151 in AOT reverse micelles. Chem. Phys. Lett. 2005, 410, 160–164. [Google Scholar] [CrossRef]
- Devji, T.; Reddy, C.; Woo, C.; Awale, S.; Kadota, S.; Carrico-Moniz, D. Pancreatic anticancer activity of a novel geranylgeranylated coumarin derivative. Bioorg. Med. Chem. Lett. 2011, 21, 5770–5773. [Google Scholar] [CrossRef]
- Reddy, N.S.; Mallireddigari, M.R.; Cosenza, S.; Gumireddy, K.; Bell, S.C.; Reddy, E.P.; Reddy, M.R. Synthesis of new coumarin 3 (N aryl) sulfonamides and their anticancer activity. Bioorg. Med. Chem. Lett. 2004, 14, 4093–4097. [Google Scholar] [CrossRef]
- Hadjipavlou-Litina, D.; Litinas, K.; Kontogiorgis, C. The Anti-inflammatory Effect of Coumarin and its Derivatives. Anti-Inflamm. Anti-Allergy Agents Med. Chem. 2007, 6, 293–306. [Google Scholar] [CrossRef]
- Xue, H.; Lu, X.; Zheng, P.; Liu, L.; Han, C.; Hu, J.; Liu, Z.; Ma, T.; Li, Y.; Wang, L.; et al. Highly Suppressing Wild-Type HIV-1 and Y181C Mutant HIV-1 Strains by 10-Chloromethyl-11-demethyl-12-oxo-calanolide A with Druggable Profile. J. Med. Chem. 2010, 53, 1397–1401. [Google Scholar] [CrossRef] [PubMed]
- Curini, M.; Epifano, F.; Maltese, F.; Marcotullio, M.C.; Gonzales, S.P.; Rodríguez, J.C. Synthesis of Collinin, an Antiviral Coumarin. Aust. J. Chem. 2003, 56, 59–60. [Google Scholar] [CrossRef]
- Kikelj, D. Peptidomimetic ThrombinInhibitors. Pathophysiol. Haemost. Thromb. 2004, 33, 487–491. [Google Scholar] [CrossRef]
- Gentilucci, L.; Tolomelli, A.; Squassabia, F. Peptides and peptidomimetics in medicine, surgery and biotechnology. Curr. Med. Chem. 2006, 13, 2449–2466. [Google Scholar] [CrossRef]
- Hwang, C.H.; Jaki, B.U.; Klein, L.L.; Lankin, D.C.; McAlpine, J.B.; Napolitano, J.G.; Fryling, N.A.; Franzblau, S.G.; Cho, S.H.; Stamets, P.E.; et al. Chlorinated Coumarins from the Polypore MushroomFomitopsis officinalisand Their Activity againstMycobacterium tuberculosis. J. Nat. Prod. 2013, 76, 1916–1922. [Google Scholar] [CrossRef] [Green Version]
- Arshad, A.; Osman, H.; Bagley, M.; Lam, C.K.; Mohamad, S.; Zahariluddin, A.S.M. Synthesis and antimicrobial properties of some new thiazolyl coumarin derivatives. Eur. J. Med. Chem. 2011, 46, 3788–3794. [Google Scholar] [CrossRef]
- Upadhyay, K.; Manvar, A.; Rawal, K.; Joshi, S.; Trivedi, J.; Chaniyara, R.; Shah, A.K. Evaluation of Structurally Diverse Benzoazepines Clubbed with Coumarins as Mycobacterium tuberculosis Agents. Chem. Boil. Drug Des. 2012, 80, 1003–1008. [Google Scholar] [CrossRef]
- Cottiglia, F.; Loy, G.; Garau, D.; Floris, C.; Casu, M.; Pompei, R.; Bonsignore, L. Antimicrobial evaluation of coumarins and flavonoids from the stems of Daphne gnidium L. Phytomedicine 2001, 8, 302–305. [Google Scholar] [CrossRef]
- Smyth, T.; Ramachandran, V.; Smyth, W. A study of the antimicrobial activity of selected naturally occurring and synthetic coumarins. Int. J. Antimicrob. Agents 2009, 33, 421–426. [Google Scholar] [CrossRef]
- Shi, Y.; Zhou, C.H. Synthesis and evaluation of a class of new coumarin triazole derivatives as potential antimicrobial agents. Bioorg. Med. Chem. Lett. 2011, 21, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Dennekamp, M.; Howarth, S.; Dick, C.A.J.; Cherrie, J.W.; Donaldson, K.; Seaton, A. Ultrafine particles and nitrogen oxides generated by gas and electric cooking. Occup. Environ. Med. 2001, 58, 511–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukasiewicz, J.; Jachymek, W.; Niedziela, T.; Dzieciatkowska, M.; Lakomska, J.; Międzybrodzki, R.; Fortuna, W.; Szymaniec, S.; Misiuk Hojlo, M.; Lugowski, C. Serological characterization of anti endotoxin serumdirected against the conjugate of oligosaccharide core of Escherichia coli type R4 with tetanus toxoid. FEMS Immunol. Med. Microbiol. 2003, 37, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Nnalue, N.A.; Khan, G.N.; Mustafa, N. Cross-reactivity between six Enterobacteriaceae complete lipopolysaccharide core chemotypes. J. Med. Microbiol. 1999, 48, 433–441. [Google Scholar] [CrossRef]
- Heinrichs, D.E.; Yethon, J.A.; Amor, P.A.; Whitfield, C. The assembly system for the outer core portion of R1 and R4 type lipopolysaccharides of Escherichia coli the r1 core specific β glucosyltransferase provides a novel attachment site for Opolysaccharides. J. Biol. Chem. 1998, 273, 29497–29505. [Google Scholar] [CrossRef] [Green Version]
- Amor, K.; Heinrichs, D.E.; Frirdich, E.; Ziebell, K.; Johnson, R.P.; Whitfield, C. Distribution of Core Oligosaccharide Types in Lipopolysaccharides from Escherichia coli. Infect. Immun. 2000, 68, 1116–1124. [Google Scholar] [CrossRef] [Green Version]
- Appelmelk, B.J.; An, Y.; Hekker, T.A.M.; Thijs, L.G.; MacLaren, D.M.; De Graaf, J. Frequencies of lipopolysaccharide core types in Escherichia coli strains from bacteraemic patients. Microbiology 1994, 140, 1119–1124. [Google Scholar] [CrossRef] [Green Version]
- Kowalczyk, P.; Borkowski, A.; Czerwonka, G.; Cłapa, T.; Cieśla, J.; Misiewicz, A.; Borowiec, M.; Szala, M. The microbial toxicity of quaternary ammonium ionic liquids is dependent on the type of lipopolysaccharide. J. Mol. Liq. 2018, 266, 540–547. [Google Scholar] [CrossRef]
- Borkowski, A.; Ławniczak, Ł.; Cłapa, T.; Narożna, D.; Selwet, M.; Pęziak, D.; Markiewicz, B.; Chrzanowski, Ł. Different antibacterial activity of novel theophylline-based ionic liquids–Growth kinetic and cytotoxicity studies. Ecotoxicol. Environ. Saf. 2016, 130, 54–64. [Google Scholar] [CrossRef]
- Madej, A.; Paprocki, D.; Koszelewski, D.; Żądło-Dobrowolska, A.; Brzozowska, A.; Walde, P.; Ostaszewski, R. Efficient Ugi reactions in an aqueous vesicle system. RSC Adv. 2017, 7, 33344. [Google Scholar] [CrossRef] [Green Version]
- Stromberg, Z.; Van Goor, A.; Redweik, G.A.J.; Brand, M.J.W.; Wannemuehler, M.J.; Mellata, M. Pathogenic and non-pathogenic Escherichia coli colonization and host inflammatory response in a defined microbiota mouse model. Dis. Model. Mech. 2018, 11, dmm035063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamoorthy, K.; Manivannan, G.; Kim, S.-J.; Jeyasubramanian, K.; Premanathan, M. Antibacterial activity of MgO nanoparticles based on lipid peroxidation by oxygen vacancy. J. Nanoparticle Res. 2012, 14, 1063. [Google Scholar] [CrossRef]
- Hough-Troutman, W.L.; Smiglak, M.; Griffin, S.; Reichert, W.M.; Mirska, I.; Jodynis-Liebert, J.; Adamska, T.; Nawrot, J.; Stasiewicz, M.; Rogers, R.D.; et al. Ionic liquids with dual biological function: Sweet and anti-microbial, hydrophobic quaternary ammonium-based salts. New J. Chem. 2009, 33, 26–33. [Google Scholar] [CrossRef]
- Inácio, Â.; Domingues, N.; Nunes, A.; Martins, P.; Moreno, M.J.; Estronca, L.; Fernandes, R.; Moreno, A.J.; Borrego, M.J.; Gomes, J.P.; et al. Quaternary ammonium surfactant structure determines selective toxicity towards bacteria: Mechanisms of action and clinical implications in antibacterial prophylaxis. J. Antimicrob. Chemother. 2015, 71, 641–654. [Google Scholar] [CrossRef] [Green Version]
- Jurado, J.; Saparbaev, M.; Matray, T.J.; Greenberg, M.M.; Laval, J. The Ring Fragmentation Product of Thymidine C5-Hydrate When Present in DNA is Repaired by theEscherichia coliFpg and Nth Proteins. Biochemistry 1998, 37, 7757–7763. [Google Scholar] [CrossRef]
- Cussac, C.; Laval, F. Reduction of the Toxicity and Mutagenicity of Aziridine in Mammalian Cells Harboring the Escherichia Coli fpg Gene. Nucleic Acids Res. 1996, 24, 1742–1746. [Google Scholar] [CrossRef] [Green Version]
- Kawase, M.; Varu, B.; Shah, A.K.; Motohashi, N.; Tani, S.; Saito, S.; Debnath, S.; Mahapatra, S.; Dastidar, S.G.; Chakrabarty, A.N. Antimicrobial Activity of New Coumarin Derivatives. Drug Res. 2001, 51, 67–71. [Google Scholar] [CrossRef]
- Laurin, P.; Ferroud, D.; Klich, M.; Dupuis-Hamelin, C.; Mauvais, P.; Lassaigne, P.; Bonnefoy, A.; Musicki, B. Synthesis and in vitro evaluation of novel highly potent coumarin inhibitors of gyrase B. Bioorg. Med. Chem. Lett. 1999, 9, 2079–2084. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Kincaid, J.F.; Cho, W.; Walters, D.E.; Krishnan, K.; Hussain, K.A.; Koo, Y.; Cho, H.; Rudall, C.; Holland, L.; et al. Potent HIV protease inhibitors incorporating high-affinity P2-ligands and (R)-(hydroxyethylamino)sulfonamide isostere. Bioorganic Med. Chem. Lett. 1998, 8, 687–690. [Google Scholar] [CrossRef]
- Zhao, H.; Neamati, N.; Hong, H.; Mazumder, A.; Wang, S.; Sunder, S.; Milne, G.W.A.; Pommier, Y.; Burke, T.R. Coumarin-Based Inhibitors of HIV Integrase1. J. Med. Chem. 1997, 40, 242–249. [Google Scholar] [CrossRef]
- Paprocki, D.; Koszelewski, D.; Walde, P.; Ostaszewski, R. Efficient Passerini reactions in an aqueous vesicle system. RSC Adv. 2015, 5, 102828. [Google Scholar] [CrossRef]
- Larry, K.L.; Tanka, S.K. Therapy of Helicobacter pylori Infections: Current Status and Future Directions. Annu. Rep. Med. Chem. 1995, 30, 151–158. [Google Scholar]






| Entry | R1 | R2 | R3 | Proposed Product (No.) | Present Product (No.) | Yield a |
|---|---|---|---|---|---|---|
| 1 | H | C11H23 | CH24-OMeC6H4 | 4 | 11-Dp-606 | 44% |
| 2 | NO2 | C11H23 | CH24-OMeC6H4 | 5 | 7-DP-595 | 38% |
| 3 | OMe | C11H23 | CH24-OMeC6H4 | 6 | 9-DP-571 | 17% |
| 4 | Me | C11H23 | CH24-OMeC6H4 | 7 | 16-DP-594 | 34% |
| 5 | H | C11H23 | Bz | 8 | 2-DP-577 | 38% |
| 6 | H | C11H23 | t-Bu | 9 | 10-DP-639 | 17% |
| 7 | H | C11H23 | Cyclohexyl | 10 | 1-DP578 | 29% |
| 8 | H | C11H23 | CH2COOEt | 11 | 6-DP-579 | 48% |
| 9 | H | C7H15 | CH24-OMeC6H4 | 12 | 5-DP-593 | 32% |
| 10 | H | C5H11 | CH24-OMeC6H4 | 13 | 4-DP-590 | 57% |
| 11 | H | Et | CH24-OMeC6H4 | 14 | 14-DP-589 | 13% |
| 12 | H | Bz | CH24-OMeC6H4 | 15 | 8-DP-591 | 72% |
| 13 | H | i−Pr | CH24-OMeC6H4 | 16 | 17-DP-592 | 49% |
| No. of Strain Samples | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | Type of Test |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K12 | - | *** | - | - | *** | ** | - | *** | - | *** | ** | * | * | *** | ** | ** | *** | MIC |
| R2 | - | *** | - | - | *** | ** | - | *** | - | *** | ** | * | * | *** | ** | ** | *** | MIC |
| R3 | - | *** | - | - | *** | ** | - | *** | - | *** | ** | * | * | *** | ** | ** | *** | MIC |
| R4 | - | *** | - | - | *** | ** | - | *** | - | *** | ** | * | * | *** | ** | ** | *** | MIC |
| K12 | - | - | ** | * | ** | *** | - | ** | - | * | - | *** | - | - | - | - | - | MBC |
| R2 | - | - | ** | * | ** | *** | - | ** | - | * | - | *** | - | - | - | - | - | MBC |
| R3 | - | - | ** | * | ** | *** | - | ** | - | * | - | *** | - | - | - | - | - | MBC |
| R4 | - | - | ** | * | ** | *** | - | ** | - | * | - | *** | - | - | - | - | - | MBC |
| K12 | - | - | - | *** | - | *** | * | - | ** | - | ** | - | ** | - | ** | - | - | MBC/MIC |
| R2 | - | - | - | *** | - | *** | * | - | ** | - | ** | - | ** | - | ** | - | - | MBC/MIC |
| R3 | - | - | - | *** | - | *** | * | - | ** | - | ** | - | ** | - | ** | - | - | MBC/MIC |
| R4 | - | - | - | *** | - | *** | * | - | ** | - | ** | - | ** | - | ** | - | - | MBC/MIC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalczyk, P.; Madej, A.; Paprocki, D.; Szymczak, M.; Ostaszewski, R. Coumarin Derivatives as New Toxic Compounds to Selected K12, R1–R4 E. coli Strains. Materials 2020, 13, 2499. https://doi.org/10.3390/ma13112499
Kowalczyk P, Madej A, Paprocki D, Szymczak M, Ostaszewski R. Coumarin Derivatives as New Toxic Compounds to Selected K12, R1–R4 E. coli Strains. Materials. 2020; 13(11):2499. https://doi.org/10.3390/ma13112499
Chicago/Turabian StyleKowalczyk, Paweł, Arleta Madej, Daniel Paprocki, Mateusz Szymczak, and Ryszard Ostaszewski. 2020. "Coumarin Derivatives as New Toxic Compounds to Selected K12, R1–R4 E. coli Strains" Materials 13, no. 11: 2499. https://doi.org/10.3390/ma13112499
APA StyleKowalczyk, P., Madej, A., Paprocki, D., Szymczak, M., & Ostaszewski, R. (2020). Coumarin Derivatives as New Toxic Compounds to Selected K12, R1–R4 E. coli Strains. Materials, 13(11), 2499. https://doi.org/10.3390/ma13112499