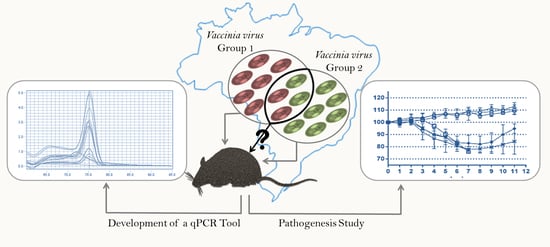
A Model to Detect Autochthonous Group 1 and 2 Brazilian Vaccinia virus Coinfections: Development of a qPCR Tool for Diagnosis and Pathogenesis Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Cells and Viruses
2.3. Animal Experiments
2.4. Plaque Phenotype
2.5. qPCR
- [ ] VACV GI = [ ] VACV total (GI+GII) − [ ] VACV GII
- [ ] VACV GI = 3.38 × 106 − 1.68 × 106
- [ ] VACV GI = 1.70 × 106
- GI = 1.70 × 106 and GII = 1.68 × 106
2.6. Statistical Analyses
3. Results
3.1. Development of qPCR Tool
3.2. A56R qPCR as a Tool to Study Pathogenesis and Viral Spread
3.2.1. Clinical Signs in Mice: Coinfected Versus Monoinfected
3.2.2. Coinfected Mice Present Higher Frequency of VACV Detection in Lungs and Spleens than Monoinfected Groups
3.2.3. Viral Load in Lung and Spleen
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Damon, I.K. Poxviruses. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams and Wilkins: Philadelphia, PA, USA, 2014; Volume 2, pp. 2160–2184. [Google Scholar]
- Moss, B. Poxviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams and Wilkins: Philadelphia, PA, USA, 2014; Volume 2, pp. 2129–2159. [Google Scholar]
- Fenner, F.; Henderson, D.A.; Arita, I.; Jezek, A.; Ladnyi, I.D. Smallpox and Its Eradication; World Health Organization Press: Geneva, Switzerland, 1988. [Google Scholar]
- Trindade, G.S.; Lobato, Z.I.; Drumond, B.P.; Leite, J.A.; Trigueiro, R.C.; Guedes, M.I.; da Fonseca, F.G.; dos Santos, J.R.; Bonjardim, C.A.; Ferreira, P.C.; et al. Short report: Isolation of two vaccinia virus strains from a single bovine vaccinia outbreak in rural area from Brazil: Implications on the emergence of zoonotic orthopoxviruses. Am. J. Trop. Med. Hyg. 2006, 75, 486–490. [Google Scholar] [PubMed]
- Singh, R.K.; Hosamani, M.; Balamurugan, V.; Bhanuprakash, V.; Rasool, T.J.; Yadav, M.P. Buffalopox: An emerging and re-emerging zoonosis. Anim. Health Res. Rev. 2007, 8, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Kroon, E.G.; Mota, B.E.; Abrahão, J.S.; da Fonseca, F.G.; de Souza Trindade, G. Zoonotic Brazilian Vaccinia virus: From field to therapy. Antivir. Res. 2011, 92, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.B.; Assis, F.L.; Ferreira, P.C.; Bonjardim, C.A.; de Souza Trindade, G.; Kroon, E.G.; Abrahão, J.S. Group 1 Vaccinia virus zoonotic outbreak in Maranhao State, Brazil. Am. J. Trop. Med. Hyg. 2013, 89, 1142–1145. [Google Scholar] [CrossRef] [PubMed]
- Franco-Luiz, A.P.; Fagundes-Pereira, A.; Costa, G.B.; Alves, P.A.; Oliveira, D.B.; Bonjardim, C.A.; Ferreira, P.C.; de Souza Trindade, G.; Panei, C.J.; Galosi, C.M.; et al. Spread of vaccinia virus to cattle herds, Argentina, 2011. Emerg. Infect. Dis. 2014, 20, 1576–1578. [Google Scholar] [CrossRef] [PubMed]
- Damaso, C.R.; Esposito, J.J.; Condit, R.C.; Moussatché, N. An emergent poxvirus from humans and cattle in Rio de Janeiro State: Cantagalo virus may derive from Brazilian smallpox vaccine. Virology 2000, 277, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Trindade, G.S.; Emerson, G.L.; Carroll, D.S.; Kroon, E.G.; Damon, I.K. Brazilian vaccinia viruses and their origins. Emerg. Infect. Dis. 2007, 13, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Megid, J.; Appolinário, C.M.; Langoni, H.; Pituco, E.M.; Okuda, L.H. Vaccinia virus in humans and cattle in southwest region of Sao Paulo state, Brazil. Am. J. Trop. Med. Hyg. 2008, 79, 647–651. [Google Scholar] [PubMed]
- Assis, F.L.; Almeida, G.M.; Oliveira, D.B.; Franco-Luiz, A.P.; Campos, R.K.; Guedes, M.I.; Fonseca, F.G.; Trindade, G.S.; Drumond, B.P.; Kroon, E.G.; et al. Characterization of a new Vaccinia virus isolate reveals the C23L gene as a putative genetic marker for autochthonous Group 1 Brazilian Vaccinia virus. PLoS ONE 2012, 7, e50413. [Google Scholar] [CrossRef] [PubMed]
- Drumond, B.P.; Leite, J.A.; da Fonseca, F.G.; Bonjardim, C.A.; Ferreira, P.C.; Kroon, E.G. Brazilian Vaccinia virus strains are genetically divergent and differ from the Lister vaccine strain. Microbes Infect. 2008, 10, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.M.; Drumond, B.P.; Guedes, M.I.; Pascoal-Xavier, M.A.; Almeida-Leite, C.M.; Arantes, R.M.; Mota, B.E.; Abrahão, J.S.; Alves, P.A.; Oliveira, F.M.; et al. Virulence in murine model shows the existence of two distinct populations of Brazilian Vaccinia virus strains. PLoS ONE 2008, 3, e3043. [Google Scholar] [CrossRef] [PubMed]
- Campos, R.K.; Brum, M.C.; Nogueira, C.E.; Drumond, B.P.; Alves, P.A.; Siqueira-Lima, L.; Assis, F.L.; Trindade, G.S.; Bonjardim, C.A.; Ferreira, P.C.; et al. Assessing the variability of Brazilian Vaccinia virus isolates from a horse exanthematic lesion: Coinfection with distinct viruses. Arch. Virol. 2011, 156, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.; Assis, F.; Almeida, G.; Albarnaz, J.; Lima, M.; Andrade, A.C.; Calixto, R.; Oliveira, C.; Diomedes Neto, J.; Trindade, G.; et al. From lesions to viral clones: Biological and molecular diversity amongst autochthonous Brazilian vaccinia virus. Viruses 2015, 7, 1218–1237. [Google Scholar] [CrossRef] [PubMed]
- Leite, J.A.; Drumond, B.P.; de Souza Trindade, G.; Bonjardim, C.A.; Ferreira, P.C.; Kroon, E.G. Brazilian Vaccinia virus strains show genetic polymorphism at the ati gene. Virus Genes 2007, 35, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Assis, F.L.; Borges, I.A.; Ferreira, P.C.; Bonjardim, C.A.; de Souza Trindade, G.; Lobato, Z.I.; Guedes, M.I.; Mesquita, V.; Kroon, E.G.; Abrahão, J.S. Group 2 vaccinia virus, Brazil. Emerg. Infect. Dis. 2012, 18, 2035–2038. [Google Scholar] [CrossRef] [PubMed]
- Brum, M.C.S.; Anjos, B.L.; Nogueira, C.E.W.; Amaral, L.A.; Weiblen, R.; Flores, E.F. An outbreak of orthopoxvirus-associated disease in horses in southern Brazil. J. Vet. Diagn. Investig. 2010, 22, 143–147. [Google Scholar]
- Trindade, G.S.; Li, Y.; Olson, V.A.; Emerson, G.; Regnery, R.L.; da Fonseca, F.G.; Kroon, E.G.; Damon, I.K. Real-time PCR assay to identify variants of Vaccinia virus: Implications for the diagnosis of bovine vaccinia in Brazil. J. Virol. Methods 2008, 152, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.L.; Wang, Y.B.; Li, M.F.; Chen, H.Y.; Guo, X.P.; Geng, J.W.; Wang, Z.Y.; Wei, Z.Y.; Cui, B.A. Simultaneous detection of porcine parvovirus and porcine circovirus type 2 by duplex real-time PCR and amplicon melting curve analysis using SYBR Green. J. Virol. Methods 2013, 187, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, G.; Balamurugan, V.; Bhanuprakash, V. TaqMan based real-time duplex PCR for simultaneous detection and quantitation of capripox and orf virus genomes in clinical samples. J. Virol. Methods 2014, 201, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; Dondi, F.; Prosperi, S.; Battilani, M. Development of a SYBR Green real-time PCR assay with melting curve analysis for simultaneous detection and differentiation of canine adenovirus type 1 and type 2. J. Virol. Methods 2015, 222, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Carletti, F.; di Caro, A.; Calcaterra, S.; Grolla, A.; Czub, M.; Ippolito, G.; Capobianchi, M.R.; Horejsh, D. Rapid, differential diagnosis of orthopox- and herpesviruses based upon real-time PCR product melting temperature and restriction enzyme analysis of amplicons. J. Virol. Methods 2005, 129, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Fedele, C.G.; Negredo, A.; Molero, F.; Sánchez-Seco, M.P.; Tenorio, A. Use of internally controlled real-time genome amplification for detection of variola virus and other orthopoxviruses infecting humans. J. Clin. Microbiol. 2006, 44, 4464–4470. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, A.; Büttner, M.; Wilhelm, S.; Pauli, G.; Meyer, H. Real-time PCR detection of parapoxvirus DNA. Clin. Chem. 2006, 52, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Olson, V.A.; Laue, T.; Laker, M.T.; Damon, I.K. Detection of monkeypox virus with real-time PCR assays. J. Clin. Virol. 2006, 36, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Abrahão, J.S.; Guedes, M.I.; Trindade, G.S.; Fonseca, F.G.; Campos, R.K.; Mota, B.F.; Lobato, Z.I.; Silva-Fernandes, A.T.; Rodrigues, G.O.; Lima, L.S.; et al. One more piece in the VACV ecological puzzle: Could peridomestic rodents be the link between wildlife and bovine vaccinia outbreaks in Brazil? PLoS ONE 2009, 4, e7428. [Google Scholar] [CrossRef] [PubMed]





| Reaction | Primers | Sequence (5’-3’) | Specificity |
|---|---|---|---|
| A | A56R-gen F | AACCACCGATGATGCGGAT | Amplify all VACV Group I and II. |
| A56R-gen R | TGCCACGGCCGACAATATAA | ||
| B | A56R-BVV-nDEL F | GCGGATCTTTATGATACGTACAATG | Amplify all VACV that do not present the 18nt deletion Group II [20]. |
| A56R-generic R | ACGGCCGACAATATAATTAATGC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calixto, R.; Oliveira, G.; Lima, M.; Andrade, A.C.; Trindade, G.D.S.; De Oliveira, D.B.; Kroon, E.G. A Model to Detect Autochthonous Group 1 and 2 Brazilian Vaccinia virus Coinfections: Development of a qPCR Tool for Diagnosis and Pathogenesis Studies. Viruses 2018, 10, 15. https://doi.org/10.3390/v10010015
Calixto R, Oliveira G, Lima M, Andrade AC, Trindade GDS, De Oliveira DB, Kroon EG. A Model to Detect Autochthonous Group 1 and 2 Brazilian Vaccinia virus Coinfections: Development of a qPCR Tool for Diagnosis and Pathogenesis Studies. Viruses. 2018; 10(1):15. https://doi.org/10.3390/v10010015
Chicago/Turabian StyleCalixto, Rafael, Graziele Oliveira, Maurício Lima, Ana Cláudia Andrade, Giliane De Souza Trindade, Danilo Bretas De Oliveira, and Erna Geessien Kroon. 2018. "A Model to Detect Autochthonous Group 1 and 2 Brazilian Vaccinia virus Coinfections: Development of a qPCR Tool for Diagnosis and Pathogenesis Studies" Viruses 10, no. 1: 15. https://doi.org/10.3390/v10010015
APA StyleCalixto, R., Oliveira, G., Lima, M., Andrade, A. C., Trindade, G. D. S., De Oliveira, D. B., & Kroon, E. G. (2018). A Model to Detect Autochthonous Group 1 and 2 Brazilian Vaccinia virus Coinfections: Development of a qPCR Tool for Diagnosis and Pathogenesis Studies. Viruses, 10(1), 15. https://doi.org/10.3390/v10010015