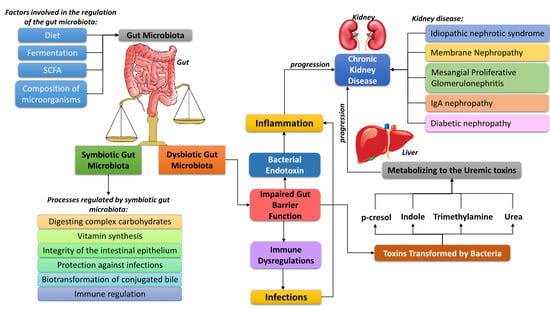
A Link between Chronic Kidney Disease and Gut Microbiota in Immunological and Nutritional Aspects
Abstract
:1. Introduction
2. The Importance of the Human Microbiota
3. The Process of Immune Modulation by Human Gut Microbiota
3.1. The Role and Importance of Bacterial Metabolites and Components in the Human Body
3.1.1. The Role of Short-Chain Fatty Acids Which Are Products of Intestinal Bacterial Metabolism in the Human Body
- In colonic epithelial cells where butyrate is the main substrate (which is the energy source for colonocytes);
- In liver cells, where the acetate produced in the gluconeogenesis process is metabolized, as well as butyrate and propionate; and
3.1.2. The Role and Importance of Indole
3.1.3. The Role and Importance of Aryl Hydrocarbon Receptor
3.1.4. The Role and Importance of Polyamines
3.2. Regulation of the Immune Response by Gut Microbiota
4. Influence of the Intestinal Microbiota on the Development of Kidney Diseases, with Particular Emphasis on the Role of the Immune System
- Reduced diversity and number of microorganisms, with a predominance of proteolytic bacteria;
- A phenomenon of the translocation of the microorganisms associated with the colonization of regions of the gastrointestinal tract that have been much less populated so far, and changes in the ratio of aerobic and anaerobic bacteria;
- The intestinal epithelial barrier is disrupted; and
4.1. Chronic Kidney Disease
4.2. Idiopathic Nephrotic Syndrome
Membrane Nephropathy and Mesangial Proliferative Glomerulonephritis
4.3. IgA Nephropathy
4.4. Diabetic Nephropathy
5. The Importance of Diet in the Progression of CKD
6. How to Restore the Symbiosis of the Gut Microbiota?
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chronic Kidney Disease Basics/Chronic Kidney Disease Initiative/CDC. Available online: https://www.cdc.gov/kidneydisease/basics.html (accessed on 11 September 2021).
- Kaufman, D.P.; Basit, H.; Knohl, S.J. Physiology, Glomerular Filtration Rate; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Filipska, A.; Bohdan, B.; Wieczorek, P.P.; Hudz, N. Chronic kidney disease and dialysis therapy: Incidence and prevalence in the world. Pharmacia 2021, 68, 463–470. [Google Scholar] [CrossRef]
- Zdrojewski, Ł.; Zdrojewski, T.; Rutkowski, M.; Bandosz, P.; Król, E.; Wyrzykowski, B.; Rutkowski, B. Prevalence of chronic kidney disease in a representative sample of the Polish population: Results of the NATPOL 2011 survey. Nephrol. Dial. Transplant. 2016, 31, 433–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobby, G.P.; Karaduta, O.; Dusio, G.F.; Singh, M.; Zybailov, B.L.; Arthur, J.M. Chronic kidney disease and the gut microbiome. Am. J. Physiol. Renal. Physiol. 2019, 316, F1211–F1217. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Fan, Y.; Li, A.; Shen, Q.; Wu, J.; Ren, L.; Lu, H.; Ding, S.; Ren, H.; Liu, C.; et al. Alterations of the human gut microbiome in chronic kidney disease. Adv. Sci. 2020, 7, 2001936. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70, S38–S44. [Google Scholar] [CrossRef] [Green Version]
- Rackaityte, E.; Lynch, S.V. The human microbiome in the 21st century. Nat. Commun. 2020, 11, 5256. [Google Scholar] [CrossRef]
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The human microbiome and its impacts on health. Int. J. Microbiol. 2020, 2020, e8045646. [Google Scholar] [CrossRef]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Duffy, A. Factors influencing the gut microbiota, inflammation, and type 2 diabetes. J. Nutr. 2017, 147, 1468S–1475S. [Google Scholar] [CrossRef] [Green Version]
- Kho, Z.Y.; Lal, S.K. The human gut microbiome—A potential controller of wellness and disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Cahana, I.; Iraqi, F.A. Impact of host genetics on gut microbiome: Take-home lessons from human and mouse studies. Anim. Models Exp. Med. 2020, 3, 229–236. [Google Scholar] [CrossRef]
- Malard, F.; Dore, J.; Gaugler, B.; Mohty, M. Introduction to host microbiome symbiosis in health and disease. Mucosal. Immunol. 2021, 14, 547–554. [Google Scholar] [CrossRef]
- Sharma, A.; Im, S.-H. Special issue on the human microbiome: From symbiosis to therapy. Exp. Mol. Med. 2020, 52, 1361–1363. [Google Scholar] [CrossRef]
- Lee, L.-H.; Wong, S.H.; Chin, S.-F.; Singh, V.; Ab Mutalib, N.-S. Editorial: Human microbiome: Symbiosis to pathogenesis. Front. Microbiol. 2021, 12, 252. [Google Scholar] [CrossRef]
- Eloe-Fadrosh, E.A.; Rasko, D.A. The human microbiome: From symbiosis to pathogenesis. Annu. Rev. Med. 2013, 64, 145–163. [Google Scholar] [CrossRef] [Green Version]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn′s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [Green Version]
- Caesar, R.; Tremaroli, V.; Kovatcheva-Datchary, P.; Cani, P.D.; Bäckhed, F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015, 22, 658–668. [Google Scholar] [CrossRef] [Green Version]
- Mao, L.; Franke, J. Symbiosis, dysbiosis, and rebiosis—The value of metaproteomics in human microbiome monitoring. Proteomics 2015, 15, 1142–1151. [Google Scholar] [CrossRef]
- Li, M.; Wang, B.; Zhang, M.; Rantalainen, M.; Wang, S.; Zhou, H.; Zhang, Y.; Shen, J.; Pang, X.; Zhang, M.; et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 2117–2122. [Google Scholar] [CrossRef] [Green Version]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition revisited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the human gut microbiota for immunomodulatory organisms. Cell 2017, 168, 928–943. [Google Scholar] [CrossRef] [Green Version]
- Lazar, V.; Ditu, L.-M.; Pircalabioru, G.G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L.; Chifiriuc, M.C. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front. Immunol. 2018, 9, 1830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (SCFAs)—Mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; van Harsselaar, J.; et al. Short chain fatty acids in human gut and metabolic health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- Jacobson, D.K.; Honap, T.P.; Ozga, A.T.; Meda, N.; Kagoné, T.S.; Carabin, H.; Spicer, P.; Tito, R.Y.; Obregon-Tito, A.J.; Reyes, L.M.; et al. Analysis of global human gut metagenomes shows that metabolic resilience potential for short-chain fatty acid production is strongly influenced by lifestyle. Sci. Rep. 2021, 11, 1724. [Google Scholar] [CrossRef]
- Rey, F.E.; Faith, J.J.; Bain, J.; Muehlbauer, M.J.; Stevens, R.D.; Newgard, C.B.; Gordon, J.I. Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 2010, 285, 22082–22090. [Google Scholar] [CrossRef] [Green Version]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.; Vadder, F.D.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [Green Version]
- Baxter, N.T.; Schmidt, A.W.; Venkataraman, A.; Kim, K.S.; Waldron, C.; Schmidt, T.M. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 2019, 10, e02566-18. [Google Scholar] [CrossRef] [Green Version]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef]
- Fellows, R.; Denizot, J.; Stellato, C.; Cuomo, A.; Jain, P.; Stoyanova, E.; Balázsi, S.; Hajnády, Z.; Liebert, A.; Kazakevych, J.; et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 2018, 9, 105. [Google Scholar] [CrossRef]
- Yuille, S.; Reichardt, N.; Panda, S.; Dunbar, H.; Mulder, I.E. Human gut bacteria as potent class I histone deacetylase inhibitors in vitro through production of butyric acid and valeric acid. PLoS ONE 2018, 13, e0201073. [Google Scholar] [CrossRef] [Green Version]
- Licciardi, P.V.; Ververis, K.; Karagiannis, T.C. Histone deacetylase inhibition and dietary short-chain fatty acids. ISRN Allergy 2011, 2011, 869647. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.P.; Karunakar, P.; Taraphder, S.; Yadav, H. Free fatty acid receptors 2 and 3 as microbial metabolite sensors to shape host health: Pharmacophysiological view. Pharmacophysiol. View. Biomed. 2020, 8, 154. [Google Scholar] [CrossRef]
- Sivaprakasam, S.; Gurav, A.; Paschall, A.V.; Coe, G.L.; Chaudhary, K.; Cai, Y.; Kolhe, R.; Martin, P.; Browning, D.; Huang, L.; et al. An essential role of Ffar2 (Gpr43) in dietary fibre-mediated promotion of healthy composition of gut microbiota and suppression of intestinal carcinogenesis. Oncogenesis 2016, 5, e238. [Google Scholar] [CrossRef] [Green Version]
- Kimura, I.; Inoue, D.; Hirano, K.; Tsujimoto, G. The SCFA receptor GPR43 and energy metabolism. Front. Endocrinol. 2014, 5, 85. [Google Scholar] [CrossRef] [Green Version]
- Kasubuchi, M.; Hasegawa, S.; Hiramatsu, T.; Ichimura, A.; Kimura, I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients 2015, 7, 2839–2849. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Kim, J.-H.; Park, B.O.; Kwak, Y.S. Perspectives on the therapeutic potential of short-chain fatty acid receptors. BMB Rep. 2014, 47, 173–178. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.H. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell Mol. Immunol. 2021, 18, 1161–1171. [Google Scholar] [CrossRef]
- Dupraz, L.; Magniez, A.; Rolhion, N.; Richard, M.L.; Da Costa, G.; Touch, S.; Mayeur, C.; Planchais, J.; Agus, A.; Danne, C.; et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal Γδ T cells. Cell Rep. 2021, 36, 109332. [Google Scholar] [CrossRef]
- Luu, M.; Visekruna, A. Short-chain fatty acids: Bacterial messengers modulating the immunometabolism of T cells. Eur. J. Immunol. 2019, 49, 842–848. [Google Scholar] [CrossRef] [Green Version]
- Bansal, T.; Alaniz, R.C.; Wood, T.K.; Jayaraman, A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 228–233. [Google Scholar] [CrossRef] [Green Version]
- Karlin, D.A.; Mastromarino, A.J.; Jones, R.D.; Stroehlein, J.R.; Lorentz, O. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J. Cancer Res. Clin. Oncol. 1985, 109, 135–141. [Google Scholar] [CrossRef]
- Zuccato, E.; Venturi, M.; Di Leo, G.; Colombo, L.; Bertolo, C.; Doldi, S.B.; Mussini, E. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Digest. Dis. Sci. 1993, 38, 514–519. [Google Scholar] [CrossRef]
- Gryp, T.; De Paepe, K.; Vanholder, R.; Kerckhof, F.-M.; Van Biesen, W.; Van de Wiele, T.; Verbeke, F.; Speeckaert, M.; Joossens, M.; Couttenye, M.M.; et al. Gut microbiota generation of protein-bound uremic toxins and related metabolites is not altered at different stages of chronic kidney disease. Kidney Int. 2020, 97, 1230–1242. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
- Shimada, Y.; Kinoshita, M.; Harada, K.; Mizutani, M.; Masahata, K.; Kayama, H.; Takeda, K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS ONE 2013, 8, e80604. [Google Scholar] [CrossRef] [Green Version]
- Satoh, M.; Hayashi, H.; Watanabe, M.; Ueda, K.; Yamato, H.; Yoshioka, T.; Motojima, M. Uremic toxins overload accelerates renal damage in a rat model of chronic renal failure. Nephron. Exp. Neurol. 2003, 95, e111–e118. [Google Scholar] [CrossRef] [PubMed]
- Brito, J.S.; Borges, N.A.; Esgalhado, M.; Magliano, D.C.; Soulage, C.O.; Mafra, D. Aryl hydrocarbon receptor activation in chronic kidney disease: Role of uremic toxins. Nephron 2017, 137, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Carlson, E.A.; McCulloch, C.; Koganti, A.; Goodwin, S.B.; Sutter, T.R.; Silkworth, J.B. Divergent transcriptomic responses to aryl hydrocarbon receptor agonists between rat and human primary hepatocytes. Toxicol. Sci. 2009, 112, 257–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safe, S.; Han, H.; Goldsby, J.; Mohankumar, K.; Chapkin, R.S. Aryl hydrocarbon receptor (AhR) ligands as selective AhR modulators: Genomic studies. Curr. Opin. Toxicol. 2018, 11, 10–20. [Google Scholar] [CrossRef]
- Feng, Y.-L.; Cao, G.; Chen, D.-Q.; Vaziri, N.D.; Chen, L.; Zhang, J.; Wang, M.; Guo, Y.; Zhao, Y.-Y. Microbiome—Metabolomics reveals gut microbiota associated with glycine-conjugated metabolites and polyamine metabolism in chronic kidney disease. Cell. Mol. Life Sci. 2019, 76, 4961–4978. [Google Scholar] [CrossRef] [Green Version]
- Ebnet, K.; Suzuki, A.; Ohno, S.; Vestweber, D. Junctional adhesion molecules (JAMs): More molecules with dual functions? J. Cell Sci. 2004, 117, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef]
- Koch, B.E.V.; Yang, S.; Lamers, G.; Stougaard, J.; Spaink, H.P. Intestinal microbiome Adjusts the innate immune setpoint during colonization through negative regulation of MyD88. Nat. Commun. 2018, 9, 4099. [Google Scholar] [CrossRef]
- Karmarkar, D.; Rock, K.L. Microbiota signalling through MyD88 is necessary for a systemic neutrophilic inflammatory response. Immunology 2013, 140, 483–492. [Google Scholar] [CrossRef]
- Schoenborn, A.A.; von Furstenberg, R.J.; Valsaraj, S.; Hussain, F.S.; Stein, M.; Shanahan, M.T.; Henning, S.J.; Gulati, A.S. The enteric microbiota regulates jejunal paneth cell number and function without impacting intestinal stem cells. Gut Microbes 2019, 10, 45–58. [Google Scholar] [CrossRef] [Green Version]
- Lueschow, S.R.; McElroy, S.J. The paneth cell: The curator and defender of the immature small intestine. Front. Immunol. 2020, 11, 587. [Google Scholar] [CrossRef]
- Peng, C.; Ouyang, Y.; Lu, N.; Li, N. The NF-ΚB signaling pathway, the microbiota, and gastrointestinal tumorigenesis: Recent advances. Front. Immunol. 2020, 11, 1387. [Google Scholar] [CrossRef]
- Lakhdari, O.; Tap, J.; Béguet-Crespel, F.; Le Roux, K.; de Wouters, T.; Cultrone, A.; Nepelska, M.; Lefèvre, F.; Doré, J.; Blottière, H.M. Identification of NF-ΚB modulation capabilities within human intestinal commensal bacteria. J. Biomed. Biotechnol. 2011, 2011, e282356. [Google Scholar] [CrossRef] [Green Version]
- Kerperien, J.; Veening-Griffioen, D.; Wehkamp, T.; van Esch, B.C.A.M.; Hofman, G.A.; Cornelissen, P.; Boon, L.; Jeurink, P.V.; Garssen, J.; Knippels, L.M.J.; et al. IL-10 receptor or TGF-β neutralization abrogates the protective effect of a specific nondigestible oligosaccharide mixture in cow-milk-allergic mice. J. Nutr. 2018, 148, 1372–1379. [Google Scholar] [CrossRef]
- Pang, X.; Tang, Y.; Ren, X.; Chen, Q.; Tang, Y.; Liang, X. Microbiota, epithelium, inflammation, and TGF-β signaling: An intricate interaction in oncogenesis. Front. Microbiol. 2018, 9, 1353. [Google Scholar] [CrossRef]
- Wan, Y.-D.; Zhu, R.-X.; Bian, Z.-Z.; Pan, X.-T. Improvement of gut microbiota by inhibition of P38 mitogen-activated protein kinase (MAPK) signaling pathway in rats with severe acute pancreatitis. Med. Sci. Monit. 2019, 25, 4609–4616. [Google Scholar] [CrossRef]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.A.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut–lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef]
- Jaglin, M.; Rhimi, M.; Philippe, C.; Pons, N.; Bruneau, A.; Goustard, B.; Daugé, V.; Maguin, E.; Naudon, L.; Rabot, S. Indole, a signaling molecule produced by the gut microbiota, negatively impacts emotional behaviors in rats. Front. Neurosci. 2018, 12, 216. [Google Scholar] [CrossRef]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef] [Green Version]
- Bermudez-Martin, P.; Becker, J.A.J.; Caramello, N.; Fernandez, S.P.; Costa-Campos, R.; Canaguier, J.; Barbosa, S.; Martinez-Gili, L.; Myridakis, A.; Dumas, M.-E.; et al. The microbial metabolite P-cresol induces autistic-like behaviors in mice by remodeling the gut microbiota. Microbiome 2021, 9, 157. [Google Scholar] [CrossRef]
- Al Hinai, E.A.; Kullamethee, P.; Rowland, I.R.; Swann, J.; Walton, G.E.; Commane, D.M. Modelling the role of microbial P-cresol in colorectal genotoxicity. Gut Microbes 2019, 10, 398–411. [Google Scholar] [CrossRef] [Green Version]
- Arias, N.; Arboleya, S.; Allison, J.; Kaliszewska, A.; Higarza, S.G.; Gueimonde, M.; Arias, J.L. The relationship between choline bioavailability from diet, intestinal microbiota composition, and its modulation of human diseases. Nutrients 2020, 12, 2340. [Google Scholar] [CrossRef]
- Romano, K.A.; Vivas, E.I.; Amador-Noguez, D.; Rey, F.E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 2015, 6, e02481-14. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Dai, M. Trimethylamine N-oxide generated by the gut microbiota is associated with vascular inflammation: New insights into atherosclerosis. Mediators Inflamm. 2020, 2020, 4634172. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Li, X.; Yang, F.; Zhao, R.; Pan, X.; Liang, J.; Tian, L.; Li, X.; Liu, L.; Xing, Y.; et al. Gut microbiota-dependent marker TMAO in promoting cardiovascular disease: Inflammation mechanism, clinical prognostic, and potential as a therapeutic target. Front. Pharmacol. 2019, 10, 1360. [Google Scholar] [CrossRef]
- Palmer, S.C.; Rabindranath, K.S.; Craig, J.C.; Roderick, P.J.; Locatelli, F.; Strippoli, G.F.M. High-flux versus low-flux membranes for end-stage kidney disease. Cochrane Database Syst. Rev. 2012, 9, CD005016. [Google Scholar] [CrossRef]
- Ichii, O.; Otsuka-Kanazawa, S.; Nakamura, T.; Ueno, M.; Kon, Y.; Chen, W.; Rosenberg, A.Z.; Kopp, J.B. Podocyte injury caused by indoxyl sulfate, a uremic toxin and aryl-hydrocarbon receptor ligand. PLoS ONE 2014, 9, e108448. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Chang, S.; Wu, M. Uremic toxins induce kidney fibrosis by activating intrarenal renin-angiotensin-aldosterone system associated epithelial-to-mesenchymal transition. PLoS ONE 2012, 7, e34026. [Google Scholar] [CrossRef] [Green Version]
- Meijers, B.K.I.; Evenepoel, P. The gut-kidney axis: Indoxyl sulfate, p-cresyl sulfate and CKD progression. Nephrol. Dial. Transplant. 2011, 26, 759–761. [Google Scholar] [CrossRef]
- Poesen, R.; Viaene, L.; Verbeke, K.; Claes, K.; Bammens, B.; Sprangers, B.; Naesens, M.; Vanrenterghem, Y.; Kuypers, D.; Evenepoel, P.; et al. Renal clearance and intestinal generation of P-cresyl sulfate and indoxyl sulfate in CKD. Clin. J. Am. Soc. Nephrol. 2013, 8, 1508–1514. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.-J.; Chuang, C.-K.; Jayakumar, T.; Liu, H.-L.; Pan, C.-F.; Wang, T.-J.; Chen, H.-H.; Wu, C.-J. Serum P-cresyl sulfate predicts cardiovascular disease and mortality in elderly hemodialysis patients. Arch. Med. Sci. 2013, 9, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; DuGar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.-M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.H.; Wang, Z.; Kennedy, D.J.; Wu, Y. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015, 116, 448–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.H.W.; Wang, Z.; Li, X.S.; Fan, Y.; Li, D.S.; Wu, Y.; Hazen, S.L. Increased trimethylamine N-oxide portends high mortality risk independent of glycemic control in patients with type 2 diabetes mellitus. Clin. Chem. 2017, 63, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Guldris, S.C.; Parra, E.G.; Amenós, A.C. Gut microbiota in chronic kidney disease. Nefrología 2017, 37, 9–19. [Google Scholar] [CrossRef]
- Stanford, J.; Charlton, K.; Stefoska-Needham, A.; Ibrahim, R.; Lambert, K. The gut microbiota profile of adults with kidney disease and kidney stones: A systematic review of the literature. BMC Nephrol. 2020, 21, 215. [Google Scholar] [CrossRef]
- Ramezani, A.; Raj, D.S. The gut microbiome, kidney disease, and targeted interventions. JASN 2014, 25, 657–670. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, M.; Wang, J.; Li, R.; Zhang, Y. Alterations to the gut microbiota and their correlation with inflammatory factors in chronic kidney disease. Front. Cell. Infect. Microbiol. 2019, 9, 206. [Google Scholar] [CrossRef] [Green Version]
- Colucci, M.; Corpetti, G.; Emma, F.; Vivarelli, M. Immunology of idiopathic nephrotic syndrome. Pediatr. Nephrol. 2018, 33, 573–584. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, D.; Lin, Z.; Zhou, W.; Rao, J.; Li, Y.; Wu, J.; Peng, H.; Lou, T. Dysbiosis of gut microbiota in adult idiopathic membranous nephropathy with nephrotic syndrome. Microb. Pathog. 2020, 147, 104359. [Google Scholar] [CrossRef]
- He, H.; Lin, M.; You, L.; Chen, T.; Liang, Z.; Li, D.; Xie, C.; Xiao, G.; Ye, P.; Kong, Y.; et al. Gut microbiota profile in adult patients with idiopathic nephrotic syndrome. BioMed Res. Int. 2021, 2021, e8854969. [Google Scholar] [CrossRef]
- Kaneko, K.; Tsuji, S.; Kimata, T. Role of gut microbiota in idiopathic nephrotic syndrome in children. Med. Hypotheses 2017, 108, 35–37. [Google Scholar] [CrossRef]
- Angelis, M.D.; Montemurno, E.; Piccolo, M.; Vannini, L.; Lauriero, G.; Maranzano, V.; Gozzi, G.; Serrazanetti, D.; Dalfino, G.; Gobbetti, M.; et al. Microbiota and metabolome associated with immunoglobulin a nephropathy (IgAN). PLoS ONE 2014, 9, e99006. [Google Scholar] [CrossRef] [Green Version]
- Dong, R.; Bai, M.; Zhao, J.; Wang, D.; Ning, X.; Sun, S. A comparative study of the gut microbiota associated with immunoglobulin a nephropathy and membranous nephropathy. Front. Cell. Infect. Microbiol. 2020, 10, 598. [Google Scholar] [CrossRef]
- Yu, W.; Shang, J.; Guo, R.; Zhang, F.; Zhang, W.; Zhang, Y.; Wu, F.; Ren, H.; Liu, C.; Xiao, J.; et al. The gut microbiome in differential diagnosis of diabetic kidney disease and membranous nephropathy. Ren. Fail. 2020, 42, 1100. [Google Scholar] [CrossRef]
- Shah, N.B.; Nigwekar, S.U.; Kalim, S.; Lelouvier, B.; Servant, F.; Dalal, M.; Krinsky, S.; Fasano, A.; Tolkoff-Rubin, N.; Allegretti, A.S. The gut and blood microbiome in IgA nephropathy and healthy controls. Kidney360 2021, 2, 1261–1274. [Google Scholar] [CrossRef]
- Coppo, R. The gut-kidney axis in IgA nephropathy: Role of microbiota and diet on genetic predisposition. Pediatr. Nephrol. 2018, 33, 53–61. [Google Scholar] [CrossRef]
- Kanbay, M.; Onal, E.M.; Afsar, B.; Dagel, T.; Yerlikaya, A.; Covic, A.; Vaziri, N.D. The crosstalk of gut microbiota and chronic kidney disease: Role of inflammation, proteinuria, hypertension, and diabetes mellitus. Int. Urol. Nephrol. 2018, 50, 1453–1466. [Google Scholar] [CrossRef] [Green Version]
- Sugurmar, A.N.K.; Mohd, R.; Shah, S.A.; Neoh, H.; Cader, R.A. Gut microbiota in immunoglobulin a nephropathy: A Malaysian perspective. BMC Nephrol. 2021, 22, 145. [Google Scholar] [CrossRef]
- Chi, M.; Ma, K.; Wang, J.; Ding, Z.; Li, Y.; Zhu, S.; Liang, X.; Zhang, Q.; Song, L.; Liu, C. The immunomodulatory effect of the gut microbiota in kidney disease. J. Immunol. Res. 2021, 2021, e5516035. [Google Scholar] [CrossRef] [PubMed]
- Al Khodor, S.; Shatat, I.F. Gut microbiome and kidney disease: A bidirectional relationship. Pediatr. Nephrol. 2017, 32, 921–931. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zhu, S.; Xu, G. Targeting gut microbiota: A potential promising therapy for diabetic kidney disease. Am. J. Transl. Res. 2016, 8, 4009–4016. [Google Scholar] [PubMed]
- Mosterd, C.M.; Kanbay, M.; van den Born, B.J.H.; van Raalte, D.H.; Rampanelli, E. Intestinal microbiota and diabetic kidney diseases: The role of microbiota and derived metabolites inmodulation of renal inflammation and disease progression. Best Pract. Res. Clin. Endocrinol. Metab. 2021, 35, 101484. [Google Scholar] [CrossRef] [PubMed]
- Thurber, C.; Dugas, L.R.; Ocobock, C.; Carlson, B.; Speakman, J.R.; Pontzer, H. Extreme events reveal an alimentary limit on sustained maximal human energy expenditure. Sci. Adv. 2019, 5, eaaw0341. [Google Scholar] [CrossRef] [Green Version]
- Johansen, K.L.; Lee, C. Body composition in chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 2015, 24, 268–275. [Google Scholar] [CrossRef] [Green Version]
- Weiner, I.D.; Mitch, W.E.; Sands, J.M. Urea and ammonia metabolism and the control of renal nitrogen excretion. Clin. J. Am. Soc. Nephrol. 2015, 10, 1444–1458. [Google Scholar] [CrossRef] [Green Version]
- Beddhu, S.; Wei, G.; Chen, X.; Boucher, R.; Kiani, R.; Raj, D.; Chonchol, M.; Greene, T.; Murtaugh, M.A. Associations of dietary protein and energy intakes with protein-energy wasting syndrome in hemodialysis patients. Kidney Int. Rep. 2017, 2, 821–830. [Google Scholar] [CrossRef] [Green Version]
- Carrero, J.J.; Stenvinkel, P.; Cuppari, L.; Ikizler, T.A.; Kalantar-Zadeh, K.; Kaysen, G.; Mitch, W.E.; Price, S.R.; Wanner, C.; Wang, A.Y.M.; et al. Etiology of the protein-energy wasting syndrome in chronic kidney disease: A consensus statement from the International Society of Renal Nutrition and Metabolism (ISRNM). J. Ren. Nutr. 2013, 23, 77–90. [Google Scholar] [CrossRef] [Green Version]
- Nutrition in CKD. Available online: www.kidney.org/professionals/guidelines/guidelines_commentaries/nutrition-ckd (accessed on 26 September 2021).
- Beto, J.A.; Ramirez, W.E.; Bansal, V.K. Medical nutrition therapy in adults with chronic kidney disease: Integrating evidence and consensus into practice for the generalist registered dietitian nutritionist. J. Acad. Nutr. Diet 2014, 114, 1077–1087. [Google Scholar] [CrossRef]
- Ikizler, T.A.; Cano, N.J.; Franch, H.; Fouque, D.; Himmelfarb, J.; Kalantar-Zadeh, K.; Kuhlmann, M.K.; Stenvinkel, P.; TerWee, P.; Teta, D.; et al. Prevention and treatment of protein energy wasting in chronic kidney disease patients: A consensus statement by the International Society of Renal Nutrition and Metabolism. Kidney Int. 2013, 84, 1096–1107. [Google Scholar] [CrossRef] [Green Version]
- James, G.; Jackson, H. European guidelines for the nutritional care of adult renal patients. EDTNA-ERCA J. 2003, 29, 23–43. [Google Scholar] [CrossRef]
- Piccoli, G.B.; Moio, M.R.; Fois, A.; Sofronie, A.; Gendrot, L.; Cabiddu, G.; D′Alessandro, C.; Cupisti, A. The diet and haemodialysis dyad: Three eras, four open questions and four paradoxes. A narrative review, towards a personalized, patient-centered approach. Nutrients 2017, 9, 372. [Google Scholar] [CrossRef] [Green Version]
- Nallu, A.; Sharma, S.; Ramezani, A.; Muralidharan, J.; Raj, D. Gut microbiome in CKD: Challenges and opportunities. Transl. Res. 2017, 179, 24–37. [Google Scholar] [CrossRef] [Green Version]
- Mafra, D.; Borges, N.; Alvarenga, L.; Esgalhado, M.; Cardozo, L.; Lindholm, B.; Stenvinkel, P. Dietary components that may influence the disturbed gut microbiota in chronic kidney disease. Nutrients 2019, 11, 496. [Google Scholar] [CrossRef] [Green Version]
- Meijers, B.; Evenepoel, P.; Anders, H.-J. Intestinal microbiome and fitness in kidney disease. Nat. Rev. Nephrol. 2019, 15, 531–545. [Google Scholar] [CrossRef]
- Montemurno, E.; Cosola, C.; Dalfino, G.; Daidone, G.; Angelis, M.D.; Gobbetti, M.; Gesualdo, L. What would you like to eat, Mr CKD microbiota? A Mediterranean diet, please! Kydney Blood Press. Res. 2014, 39, 114–123. [Google Scholar] [CrossRef]
- Ale, E.C.; Binetti, A.G. Role of probiotics, prebiotics, and synbiotics in the elderly: Insights into their applications. Front. Microbiol. 2021, 12, 19. [Google Scholar] [CrossRef]
- Duque, A.L.R.F.; Demarqui, F.M.; Santoni, M.M.; Zanelli, C.F.; Adorno, M.A.T.; Milenkovic, D.; Mesa, V.; Sivieri, K. Effect of probiotic, prebiotic, and synbiotic on the gut microbiota of autistic children using an in vitro gut microbiome model. Food Res. Int. 2021, 149, 110657. [Google Scholar] [CrossRef]
- Edwards, P.T.; Kashyap, P.C.; Preidis, G.A. Microbiota on biotics: Probiotics, prebiotics, and synbiotics to optimize growth and metabolism. Am. J. Physiol. Gastrointest. Liv. Physiol. 2020, 319, G382–G390. [Google Scholar] [CrossRef]
- Turroni, S.; Magnani, M.; Kc, P.; Lesnik, P.; Vidal, H.; Heer, M. Gut microbiome and space travelers′ health: State of the art and possible pro/prebiotic strategies for long-term space missions. Front. Physiol. 2020, 11, 1135. [Google Scholar] [CrossRef]
- Ranganathan, N.; Ranganathan, P.; Friedman, E.A.; Joseph, A.; Delano, B.; Goldfarb, D.S.; Tam, P.; Rao, A.V.; Anteyi, E.; Musso, C.G. Pilot study of probiotic dietary supplementation for promoting healthy kidney function in patients with chronic kidney disease. Adv. Ther. 2010, 27, 634–647. [Google Scholar] [CrossRef]
- Alatriste, P.V.; Urbina Arronte, R.; Gómez Espinosa, C.O.; Espinosa Cuevas, M.d.l.Á. Effect of probiotics on human blood urea levels in patients with chronic renal failure. Nutr. Hosp. 2014, 29, 582–590. [Google Scholar] [CrossRef]
- Wang, I.-K.; Wu, Y.-Y.; Yang, Y.-F.; Ting, I.-W.; Lin, C.-C.; Yen, T.-H.; Chen, J.-H.; Wang, C.-H.; Huang, C.-C.; Lin, H.-C. The effect of probiotics on serum levels of cytokine and endotoxin in peritoneal dialysis patients: A randomised, double-blind, placebo-controlled trial. Benef. Microbes 2015, 6, 423–430. [Google Scholar] [CrossRef]
- Sirich, T.L.; Plummer, N.S.; Gardner, C.D.; Hostetter, T.H.; Meyer, T.W. Effect of increasing dietary fiber on plasma levels of colon-derived solutes in hemodialysis patients. Clin. J. Am. Soc. Nephrol. 2014, 9, 1603–1610. [Google Scholar] [CrossRef] [Green Version]
- Salmean, Y.A.; Segal, M.S.; Palii, S.P.; Dahl, W.J. Fiber supplementation lowers plasma P-cresol in chronic kidney disease patients. J. Ren. Nutr. 2015, 25, 316–320. [Google Scholar] [CrossRef] [Green Version]
- Meijers, B.K.I.; De Preter, V.; Verbeke, K.; Vanrenterghem, Y.; Evenepoel, P. P-cresyl sulfate serum concentrations in haemodialysis patients are reduced by the prebiotic oligofructose-enriched inulin. Nephrol. Dial. Transplant. 2010, 25, 219–224. [Google Scholar] [CrossRef] [Green Version]
- Poesen, R.; Evenepoel, P.; de Loor, H.; Delcour, J.A.; Courtin, C.M.; Kuypers, D.; Augustijns, P.; Verbeke, K.; Meijers, B. The influence of prebiotic arabinoxylan oligosaccharides on microbiota derived uremic retention solutes in patients with chronic kidney disease: A randomized controlled trial. PLoS ONE 2016, 11, e0153893. [Google Scholar] [CrossRef] [PubMed]
- Tayebi Khosroshahi, H.; Vaziri, N.D.; Abedi, B.; Asl, B.H.; Ghojazadeh, M.; Jing, W.; Vatankhah, A.M. Effect of high amylose resistant starch (HAM-RS2) supplementation on biomarkers of inflammation and oxidative stress in hemodialysis patients: A randomized clinical trial. Hemodial. Int. 2018, 22, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Schulman, G.; Vanholder, R.; Niwa, T. AST-120 for the management of progression of chronic kidney disease. Int. J. Nephrol. Renovasc. Dis. 2014, 7, 49–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakada, Y.; Onoue, K.; Nakano, T.; Ishihara, S.; Kumazawa, T.; Nakagawa, H.; Ueda, T.; Nishida, T.; Soeda, T.; Okayama, S.; et al. AST-120, an oral carbon absorbent, protects against the progression of atherosclerosis in a mouse chronic renal failure model by preserving SFlt-1 expression levels. Sci. Rep. 2019, 9, 15571. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, J.; Tanaka, T.; Inagi, R. Effect of AST-120 in chronic kidney disease treatment: Still a controversy? Nephron 2017, 135, 201–206. [Google Scholar] [CrossRef]
- Sirisinha, S. The potential impact of gut microbiota on your health: Current status and future challenges. Asian Pac. J. Allergy Immunol. 2016, 34, 249–264. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of machine learning in human microbiome studies: A review on feature selection, biomarker identification, disease prediction and treatment. Front. Microbiol. 2021, 12, 313. [Google Scholar] [CrossRef]
- Wang, Z.-K.; Yang, Y.-S.; Chen, Y.; Yuan, J.; Sun, G.; Peng, L.-H. Intestinal microbiota pathogenesis and fecal microbiota transplantation for inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 14805–14820. [Google Scholar] [CrossRef]
- Burrello, C.; Garavaglia, F.; Cribiù, F.M.; Ercoli, G.; Lopez, G.; Troisi, J.; Colucci, A.; Guglietta, S.; Carloni, S.; Guglielmetti, S.; et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat. Commun. 2018, 9, 5184. [Google Scholar] [CrossRef]
- Aucella, F.; Valente, G.L.; Catizone, L. The role of physical activity in the CKD setting. Kidney Blood Press. Res. 2014, 39, 97–106. [Google Scholar] [CrossRef]
- Yacoub, R.; Habib, H.; Lahdo, A.; Al Ali, R.; Varjabedian, L.; Atalla, G.; Kassis Akl, N.; Aldakheel, S.; Alahdab, S.; Albitar, S. Association between smoking and chronic kidney disease: A case control study. BMC Public Health 2010, 10, 731. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wang, L.; Ma, Z.; Zhong, L.; Wang, Y.; Gao, Y.; He, L.; Su, X. Cigarette smoking and chronic kidney disease in the general population: A systematic review and meta-analysis of prospective cohort studies. Nephrol. Dial. Transplant. 2017, 32, 475–487. [Google Scholar] [CrossRef]



| Enterotype Name | Microbiological Diversity | The Main Source of Energy | Production of Vitamins | Diet or Diet Components |
|---|---|---|---|---|
| Enterotype I | The most common bacteria are Bacteroides spp. | sugars and protein fermentation | biotin, riboflavin, panthenol, ascorbic acid and thiamine. | meat and products such as mayonnaise, cream, cheese, and other products containing large amounts of saturated fat |
| Enterotype II | The most common bacteria are Prevotella spp. | high ability to break down glycoproteins, especially mucins | biotin, riboflavin, panthenol, ascorbic acid and thiamine. | simple sugars and in vegetarians, Mediterranean ones, rich in fruits and vegetables |
| Enterotype III | The most common bacteria are Ruminococcus and Akkermansia spp. | protein fermentation, decomposition of mucin and simple sugars | biotin, riboflavin, panthenol, ascorbic acid and thiamine, folic acid | excess of alcohol and products rich in polyunsaturated fatty acids |
| Compound | Source in the Diet | Compound by Microorganisms Transformation | Compound by Liver Metabolism | Health Consequences |
|---|---|---|---|---|
| Tyrosine | Turkey, chicken, beef, brown rice, fish, milk, nuts, yogurt, eggs, cheese, fruit and vegetables | p-cresol | p-cresol sulfate | Increased gene expression associated with tubular interstitial fibrosis, aorta and vascular calcification, endothelial cell damage. It lowers the production of erythropoietin and bone rotation |
| Tryptophan | Beef, poultry, pork, fish, milk, yoghurt, eggs, soy products | Indole | p-indoxyl sulfate | Renal fibrosis, oxidative stress, increased inflammation cytokines, mortality. Braking endothelial proliferation, increased endothelial permeability. |
| Phosphatidylcholine and choline | fish and seafood, meat and dairy products | Trimethylamine | Trimethylamine N-oxide (TMNO) | Associated with higher mortality |
| Protein and nitrogen compounds | Dairy products, eggs | Urea | Ammonia | Damage to intestinal epithelial cells due to an increase in the pH of the intestinal environment |
| Features | Probiotics | Prebiotics | Synbiotics |
|---|---|---|---|
| Examples of microorganisms | Lactobacillus spp., Streptococcus spp., Saccharomyces spp., Aspergillus spp. | - | Lactobacillus rhamnosus, Bifidobacterium lactis |
| Diet ingredients rich in these microorganisms or compounds | They are found mainly in fermented products dairy, pickled vegetables and fruits, fermented sausages, sourdough cakes, sauerkraut, beer, wine and food silage as well as pharmaceutical preparations | Natural: of plant origin, including in garlic (9–16%), chicory (13–20%), artichokes (15–20%), asparagus (10–15%), onions (2–6%), wheat (1–4%) and bananas (0.3–0.7%) Artificial: lactulose, galacto-oligosaccharides, fructo-oligosaccharides, malotoligosaccharides, cyclodextrins, lactosucrose | Pharmaceutical preparations which contain selected strains of bacteria with additives that facilitate colonization, such as inulin or bean fibers, fermented milk drinks |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mertowska, P.; Mertowski, S.; Wojnicka, J.; Korona-Głowniak, I.; Grywalska, E.; Błażewicz, A.; Załuska, W. A Link between Chronic Kidney Disease and Gut Microbiota in Immunological and Nutritional Aspects. Nutrients 2021, 13, 3637. https://doi.org/10.3390/nu13103637
Mertowska P, Mertowski S, Wojnicka J, Korona-Głowniak I, Grywalska E, Błażewicz A, Załuska W. A Link between Chronic Kidney Disease and Gut Microbiota in Immunological and Nutritional Aspects. Nutrients. 2021; 13(10):3637. https://doi.org/10.3390/nu13103637
Chicago/Turabian StyleMertowska, Paulina, Sebastian Mertowski, Julia Wojnicka, Izabela Korona-Głowniak, Ewelina Grywalska, Anna Błażewicz, and Wojciech Załuska. 2021. "A Link between Chronic Kidney Disease and Gut Microbiota in Immunological and Nutritional Aspects" Nutrients 13, no. 10: 3637. https://doi.org/10.3390/nu13103637
APA StyleMertowska, P., Mertowski, S., Wojnicka, J., Korona-Głowniak, I., Grywalska, E., Błażewicz, A., & Załuska, W. (2021). A Link between Chronic Kidney Disease and Gut Microbiota in Immunological and Nutritional Aspects. Nutrients, 13(10), 3637. https://doi.org/10.3390/nu13103637