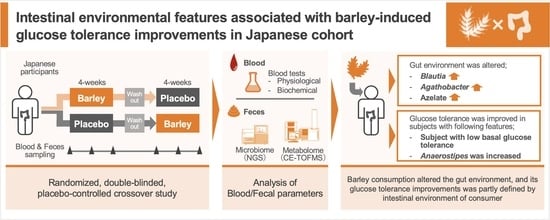
Metabologenomic Approach Reveals Intestinal Environmental Features Associated with Barley-Induced Glucose Tolerance Improvements in Japanese: A Randomized Controlled Trial
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Trial Design and Recruitment
2.3. Trial Intervention: Randomization and Blinding
2.4. DNA Extraction and 16S rRNA Gene-Based Microbiome Analysis
2.5. Metabolite Extraction and CE-TOFMS-Based Metabolome Analysis
2.6. Bioinformatics and Statistical Analysis
2.7. Defining Responders with Specific Response
3. Results
3.1. The Effect of Barley Intake on Primary and Secondary Outcomes
3.2. Effect of Barley Intake on Intestinal Microbiome and Metabolome Profiles
3.3. Characteristics of Barley Responders with Glucose Tolerance Improvement
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nilsson, A.; Östman, E.; Preston, T.; Björck, I. Effects of GI vs content of cereal fibre of the evening meal on glucose tolerance at a subsequent standardized breakfast. Eur. J. Clin. Nutr. 2008, 62, 712–720. [Google Scholar] [CrossRef]
- Bodinham, C.L.; Smith, L.; Thomas, E.L.; Bell, J.D.; Swann, J.R.; Costabile, A.; Russell-Jones, D.; Umpleby, A.M.; Robertson, M.D. Efficacy of increased resistant starch consumption in human type 2 diabetes. Endocr. Connect. 2014, 3, 75–84. [Google Scholar] [CrossRef]
- Behall, K.M.; Scholfield, D.J.; Hallfrisch, J. Diets containing barley significantly reduce lipids in mildly hypercholesterolemic men and women. Am. J. Clin. Nutr. 2004, 80, 1185–1193. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.; Granfeldt, Y.; Östman, E.; Preston, T.; Björck, I. Effects of GI and content of indigestible carbohydrates of cereal-based evening meals on glucose tolerance at a subsequent standardised breakfast. Eur. J. Clin. Nutr. 2006, 60, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, G.H.; Whyte, J.; McArthur, R.; Nestel, P.J. Barley and wheat foods: Influence on plasma cholesterol concentrations in hypercholesterolemic men. Am. J. Clin. Nutr. 1991, 53, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef]
- Yabe, D.; Seino, Y.; Fukushima, M.; Seino, S. β Cell Dysfunction Versus Insulin Resistance in the Pathogenesis of Type 2 Diabetes in East Asians. Curr. Diabetes Rep. 2015, 15, 36. [Google Scholar] [CrossRef]
- Tett, A.; Huang, K.D.; Asnicar, F.; Fehlner-Peach, H.; Pasolli, E.; Karcher, N.; Armanini, F.; Manghi, P.; Bonham, K.; Zolfo, M.; et al. The Prevotella copri Complex Comprises Four Distinct Clades Underrepresented in Westernized Populations. Cell Host Microbe 2019, 26, 666–679. [Google Scholar] [CrossRef]
- Nishijima, S.; Suda, W.; Oshima, K.; Kim, S.W.; Hirose, Y.; Morita, H.; Hattori, M. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016, 23, 125–133. [Google Scholar] [CrossRef]
- Murakami, S.; Goto, Y.; Ito, K.; Hayasaka, S.; Kurihara, S.; Soga, T.; Tomita, M.; Fukuda, S. The Consumption of Bicarbonate-Rich Mineral Water Improves Glycemic Control. Evid.-Based Complement. Altern. Med. 2015, 2015, 824395. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.W.; Suda, W.; Kim, S.; Oshima, K.; Fukuda, S.; Ohno, H.; Morita, H.; Hattori, M. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 2013, 20, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-G.; Sakamoto, K.; Seo, S.-U.; Pickard, J.M.; Gillilland, M.G.; Pudlo, N.A.; Hoostal, M.; Li, X.; Wang, T.D.; Feehley, T.; et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 2017, 356, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Martínez, I.; Lattimer, J.M.; Hubach, K.L.; Case, J.A.; Yang, J.; Weber, C.G.; Louk, J.A.; Rose, D.J.; Kyureghian, G.; Peterson, D.A.; et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013, 7, 269–280. [Google Scholar] [CrossRef]
- Ozato, N.; Saito, S.; Yamaguchi, T.; Katashima, M.; Tokuda, I.; Sawada, K.; Katsuragi, Y.; Kakuta, M.; Imoto, S.; Ihara, K.; et al. Blautia genus associated with visceral fat accumulation in adults 20–76 years of age. NPJ Biofilms Microbiomes 2019, 5, 28. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Murri, M.; Leiva, I.; Gomez-Zumaquero, J.M.; Tinahones, F.J.; Cardona, F.; Soriguer, F.; Queipo-Ortuño, M.I. Gut microbiota in children with type 1 diabetes differs from that in healthy children: A case-control study. BMC Med. 2013, 11, 46. [Google Scholar] [CrossRef]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in Gut Microbiota in Patients With vs Without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology 2020, 158, 930–946. [Google Scholar] [CrossRef]
- Karcher, N.; Pasolli, E.; Asnicar, F.; Huang, K.D.; Huang, K.D.; Tett, A.; Manara, S.; Armanini, F.; Bain, D.; Duncan, S.H.; et al. Analysis of 1321 Eubacterium rectale genomes from metagenomes uncovers complex phylogeographic population structure and subspecies functional adaptations. Genome Biol. 2020, 21, 138. [Google Scholar] [CrossRef] [PubMed]
- Millet, S.; van Oeckel, M.J.; Aluwé, M.; Delezie, E.; de Brabander, D.L. Prediction of in vivo short-chain fatty acid production in hindgut fermenting mammals: Problems and pitfalls. Crit. Rev. Food Sci. Nutr. 2010, 50, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Muthulakshmi, S.; Saravanan, R. Efficacy of azelaic acid on hepatic key enzymes of carbohydrate metabolism in high fat diet induced type 2 diabetic mice. Biochimie 2013, 95, 1239–1244. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; Molinaro, A.; Ståhlman, M.; Khan, M.T.; Schmidt, C.; Mannerås-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018, 175, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Utzschneider, K.; Neuhouser, M.L.; Newton, K.M.; Breymeyer, K.; Hullar, M.A. 229-OR: The Gut Microbiome in Adults with Prediabetes and Effect of Dietary Glycemic Index. Diabetes 2020, 69, 229-OR. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Godneva, A.; Bar, N.; Kurilshikov, A.; Lotan-Pompan, M.; Weinberger, A.; Fu, J.; Wijmenga, C.; Zhernakova, A.; et al. Structural variation in the gut microbiome associates with host health. Nature 2019, 568, 43–48. [Google Scholar] [CrossRef]
- Allen-Vercoe, E.; Daigneault, M.; White, A.; Panaccione, R.; Duncan, S.H.; Flint, H.J.; O’Neal, L.; Lawson, P.A. Anaerostipes hadrus comb. nov., a dominant species within the human colonic microbiota; reclassification of Eubacterium hadrum Moore et al. 1976. Anaerobe 2012, 18, 523–529. [Google Scholar] [CrossRef]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Shi, Y.; Shen, Q.; Dong, W.; Liu, R.; Ling, Y.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 1156, 1151–1156. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Sonnenburg, J.L. Starving our Microbial Self: The Deleterious Consequences of a Diet Deficient in Microbiota-Accessible Carbohydrates. Cell Metab. 2014, 20, 779–786. [Google Scholar] [CrossRef]




| C2 vs. T2 | C3 vs. T3 | |||
|---|---|---|---|---|
| Effect Size (95% CI) *1 | p-Value *2 | Effect Size (95% CI) *1 | p-Value *2 | |
| Blood glucose AUC (0–120 min) | −0.019 (−0.461 to 0.424) | 0.934 | 0.268 (−0.180 to 0.723) | 0.238 |
| Insulin AUC (0–120 min) | 0.018 (−0.425 to 0.460) | 0.937 | −0.123 (−0.569 to 0.320) | 0.582 |
| Blood glucose iAUC (0–120 min) | −0.099 (−0.544 to 0.343) | 0.657 | 0.258 (−0.189 to 0.712) | 0.256 |
| Insulin iAUC (0–120 min) | 0.018 (−0.425 to 0.460) | 0.937 | −0.091 (−0.536 to 0.351) | 0.682 |
| Fasting blood glucose | 0.241 (−0.205 to 0.694) | 0.287 | 0.021 (−0.421 to 0.464) | 0.924 |
| Fasting blood insulin | 0.009 (−0.434 to 0.451) | 0.968 | −0.277 (−0.733 to 0.171) | 0.223 |
| Stool frequency | 0.122 (−0.321 to 0.568) | 0.585 | 0.267 (−0.181 to 0.722) | 0.240 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goto, Y.; Nishimoto, Y.; Murakami, S.; Nomaguchi, T.; Mori, Y.; Ito, M.; Nakaguro, R.; Kudo, T.; Matsuoka, T.; Yamada, T.; et al. Metabologenomic Approach Reveals Intestinal Environmental Features Associated with Barley-Induced Glucose Tolerance Improvements in Japanese: A Randomized Controlled Trial. Nutrients 2022, 14, 3468. https://doi.org/10.3390/nu14173468
Goto Y, Nishimoto Y, Murakami S, Nomaguchi T, Mori Y, Ito M, Nakaguro R, Kudo T, Matsuoka T, Yamada T, et al. Metabologenomic Approach Reveals Intestinal Environmental Features Associated with Barley-Induced Glucose Tolerance Improvements in Japanese: A Randomized Controlled Trial. Nutrients. 2022; 14(17):3468. https://doi.org/10.3390/nu14173468
Chicago/Turabian StyleGoto, Yuka, Yuichiro Nishimoto, Shinnosuke Murakami, Tatsuhiro Nomaguchi, Yuka Mori, Masaki Ito, Ryohei Nakaguro, Toru Kudo, Tsubasa Matsuoka, Takuji Yamada, and et al. 2022. "Metabologenomic Approach Reveals Intestinal Environmental Features Associated with Barley-Induced Glucose Tolerance Improvements in Japanese: A Randomized Controlled Trial" Nutrients 14, no. 17: 3468. https://doi.org/10.3390/nu14173468
APA StyleGoto, Y., Nishimoto, Y., Murakami, S., Nomaguchi, T., Mori, Y., Ito, M., Nakaguro, R., Kudo, T., Matsuoka, T., Yamada, T., Kobayashi, T., & Fukuda, S. (2022). Metabologenomic Approach Reveals Intestinal Environmental Features Associated with Barley-Induced Glucose Tolerance Improvements in Japanese: A Randomized Controlled Trial. Nutrients, 14(17), 3468. https://doi.org/10.3390/nu14173468