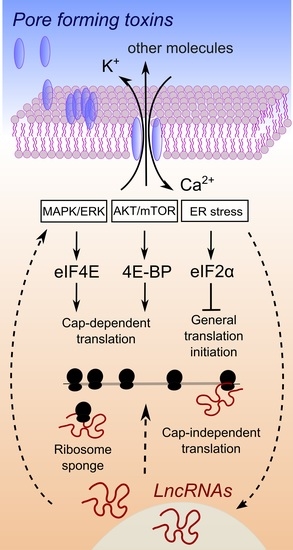
The Unexpected Tuners: Are LncRNAs Regulating Host Translation during Infections?
Abstract
:1. Membrane Damaging Toxins, Osmotic Imbalance and Translation
2. Host Long Non-Coding RNAs (LncRNAs): An Overlooked Toolkit for Controlling Gene Expression in Host–Pathogen Interaction Studies
3. Are LncRNAs Overlooked Translation Regulators in Host–Pathogen Crosstalk?
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Schiavo, G.; van der Goot, F.G. The bacterial toxin toolkit. Nat. Rev. Mol. Cell Biol. 2001, 2, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Dal Peraro, M.; van der Goot, F.G. Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 2016, 14, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Bahnan, W.; Wiley, D.J.; Barber, G.; Fields, K.A.; Schesser, K. Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion. J. Biol. Chem. 2012, 287, 28738–28744. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.P.; Topisirovic, I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Menestrina, G.; Dalla Serra, M.; Comai, M.; Coraiola, M.; Viero, G.; Werner, S.; Colin, D.A.; Monteil, H.; Prévost, G. Ion channels and bacterial infection: The case of beta-barrel pore-forming protein toxins of Staphylococcus aureus. FEBS Lett. 2003, 552, 54–60. [Google Scholar] [CrossRef]
- Kao, C.-Y.; Los, F.C.O.; Huffman, D.L.; Wachi, S.; Kloft, N.; Husmann, M.; Karabrahimi, V.; Schwartz, J.-L.; Bellier, A.; Ha, C.; et al. Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 2011, 7, e1001314. [Google Scholar] [CrossRef] [PubMed]
- Menestrina, G.; Dalla Serra, M.; Prévost, G. Mode of action of beta-barrel pore-forming toxins of the staphylococcal gamma-hemolysin family. Toxicon 2001, 39, 1661–1672. [Google Scholar] [CrossRef]
- Kloft, N.; Busch, T.; Neukirch, C.; Weis, S.; Boukhallouk, F.; Bobkiewicz, W.; Cibis, I.; Bhakdi, S.; Husmann, M. Pore-forming toxins activate MAPK p38 by causing loss of cellular potassium. Biochem. Biophys. Res. Commun. 2009, 385, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Wolfmeier, H.; Schoenauer, R.; Atanassoff, A.P.; Neill, D.R.; Kadioglu, A.; Draeger, A.; Babiychuk, E.B. Ca2+-dependent repair of pneumolysin pores: A new paradigm for host cellular defense against bacterial pore-forming toxins. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 2045–2054. [Google Scholar] [CrossRef] [PubMed]
- Bischofberger, M.; Iacovache, I.; Gisou van der Goot, F. Pathogenic Pore-Forming Proteins: Function and Host Response. Cell Host Microbe 2012, 12, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Los, F.C.O.; Randis, T.M.; Aroian, R.V.; Ratner, A.J. Role of Pore-Forming Toxins in Bacterial Infectious Diseases. Microbiol. Mol. Biol. Rev. 2013, 77, 173–207. [Google Scholar] [CrossRef] [PubMed]
- Meixenberger, K.; Pache, F.; Eitel, J.; Schmeck, B.; Hippenstiel, S.; Slevogt, H.; N’Guessan, P.; Witzenrath, M.; Netea, M.G.; Chakraborty, T.; et al. Listeria monocytogenes-Infected Human Peripheral Blood Mononuclear Cells Produce IL-1, Depending on Listeriolysin O and NLRP3. J. Immunol. 2010, 184, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Clamer, M.; Tebaldi, T.; Marchioretto, M.; Bernabo, P.; Bertini, E.; Guella, G.; Serra, M.D.; Quattrone, A.; Viero, G. Global translation variations in host cells upon attack of lytic and sublytic Staphylococcus aureus-haemolysin. Biochem. J. 2015, 472, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Chong, P.A.; Forman-Kay, J.D. Liquid-liquid phase separation in cellular signaling systems. Curr. Opin. Struct. Biol. 2016, 41, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Banjade, S.; Cheng, H.-C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, R.J.C.; Serra, M.D.; Froelich, C.J.; Wallace, M.I.; Anderluh, G. Membrane pore formation at protein-lipid interfaces. Trends Biochem. Sci. 2014, 39, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.R.; Bischofberger, M.; Frêche, B.; Ho, S.; Parton, R.G.; van der Goot, F.G. Pore-forming toxins induce multiple cellular responses promoting survival. Cell. Microbiol. 2011, 13, 1026–1043. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.T.; McNeil, P.L. Membrane Repair: Mechanisms and Pathophysiology. Physiol. Rev. 2015, 95, 1205–1240. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, A.; Awad, M.M.; Cheung, J.K.; Hiscox, T.J.; Lyras, D.; Rood, J.I. The pore-forming α-toxin from clostridium septicum activates the MAPK pathway in a Ras-c-Raf-dependent and independent manner. Toxins 2015, 7, 516–534. [Google Scholar] [CrossRef] [PubMed]
- Iordanov, M.S.; Magun, B.E. Loss of cellular K+ mimics ribotoxic stress. Inhibition of protein synthesis and activation of the stress kinases SEK1/MKK4, stress-activated protein kinase/c-Jun NH2-terminal kinase 1, and p38/HOG1 by palytoxin. J. Biol. Chem. 1998, 273, 3528–3534. [Google Scholar] [CrossRef] [PubMed]
- Tawk, M.Y.; Zimmermann-Meisse, G.; Bossu, J.L.; Potrich, C.; Bourcier, T.; Dalla Serra, M.; Poulain, B.; Prévost, G.; Jover, E. Internalization of staphylococcal leukotoxins that bind and divert the C5a receptor is required for intracellular Ca2+ mobilization by human neutrophils. Cell. Microbiol. 2015, 17, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Barry, K.C.; Ingolia, N.T.; Vance, R.E. Global analysis of gene expression reveals mRNA superinduction is required for the inducible immune response to a bacterial pathogen. eLife 2017, 6, e22707. [Google Scholar] [CrossRef] [PubMed]
- Tiedje, C.; Diaz-Muñoz, M.D.; Trulley, P.; Ahlfors, H.; Laaß, K.; Blackshear, P.J.; Turner, M.; Gaestel, M. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. 2016, 44, 7418–7440. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, M.; Rooney, M.S.; Mertins, P.; Przybylski, D.; Chevrier, N.; Satija, R.; Rodriguez, E.H.; Fields, A.P.; Schwartz, S.; Raychowdhury, R.; et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science 2015, 347, 1259038. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Pan, X.; Zhou, H.-R.; Pestka, J.J. Modulation of Inflammatory Gene Expression by the Ribotoxin Deoxynivalenol Involves Coordinate Regulation of the Transcriptome and Translatome. Toxicol. Sci. 2013, 131, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Ceppi, M.; Clavarino, G.; Gatti, E.; Schmidt, E.K.; de Gassart, A.; Blankenship, D.; Ogola, G.; Banchereau, J.; Chaussabel, D.; Pierre, P. Ribosomal protein mRNAs are translationally-regulated during human dendritic cells activation by LPS. Immun. Res. 2009, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, H.; Ito, M.; Yuasa, T.; Kikuguchi, C.; Hijikata, A.; Takayama, M.; Kimura, Y.; Yokoyama, R.; Kaji, T.; Ohara, O. Genome-wide identification and characterization of transcripts translationally regulated by bacterial lipopolysaccharide in macrophage-like J774.1 cells. Physiol. Genom. 2008, 33, 121–132. [Google Scholar] [CrossRef] [PubMed]
- GENCODE. Available online: https://www.gencodegenes.org/ (accessed on 10 October 2017).
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.-K.; Koche, R.P.; et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Arthanari, Y.; Heintzen, C.; Griffiths-Jones, S.; Crosthwaite, S.K. Natural antisense transcripts and long non-coding RNA in Neurospora crassa. PLoS ONE 2014, 9, e91353. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, D.; Zhang, W.; Guo, M.; Zhan, Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012, 31, 4415–4427. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.-J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Milligan, M.J.; Harvey, E.; Yu, A.; Morgan, A.L.; Smith, D.L.; Zhang, E.; Berengut, J.; Sivananthan, J.; Subramaniam, R.; Skoric, A.; et al. Global Intersection of Long Non-Coding RNAs with Processed and Unprocessed Pseudogenes in the Human Genome. Front. Genet. 2016, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Schein, A.; Zucchelli, S.; Kauppinen, S.; Gustincich, S.; Carninci, P. Identification of antisense long noncoding RNAs that function as SINEUPs in human cells. Sci. Rep. 2016, 6, 33605. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; St. Laurent, G.; Raz, T.; Ozsolak, F.; Reynolds, C.P.; Sorensen, P.H. B.; Reaman, G.; Milos, P.; Arceci, R.J.; Thompson, J.F.; et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is “dark matter” un-annotated RNA. BMC Biol. 2010, 8, 149. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Ribelles, M.; Solé, C.; Xu, Z.; Steinmetz, L.M.; de Nadal, E.; Posas, F. Control of Cdc28 CDK1 by a stress-induced lncRNA. Mol. Cell 2014, 53, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, H.; Mei, Y.; Wu, M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol. Cell 2014, 53, 88–100. [Google Scholar] [CrossRef] [PubMed]
- St. Laurent, G.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef]
- Niazi, F.; Valadkhan, S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA 2012, 18, 825–843. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, S.; Liu, X.; Liu, H.; Hu, T.; Qiu, X.; Zhang, J.; Lei, M. Analyses of Long Non-Coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 2016, 6, 20238. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.A.; Zhang, M.; Huang, J.; Modarresi, F.; Van der Brug, M.P.; Nalls, M.A.; Cookson, M.R.; St. Laurent, G.; Wahlestedt, C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010, 11, R56. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Van Heesch, S.; van Iterson, M.; Jacobi, J.; Boymans, S.; Essers, P.B.; de Bruijn, E.; Hao, W.; MacInnes, A.W.; Cuppen, E.; Simonis, M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014, 15, R6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlevaro-Fita, J.; Rahim, A.; Guigó, R.; Vardy, L.A.; Johnson, R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 2016, 22, 867–882. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.A.; Alba, M.M. Long non-coding RNAs as a source of new peptides. eLife 2014, 3, e03523. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.-T.; Su, H.; Khodadadi-Jamayran, A.; Lin, S.; Zhang, L.; Zhou, D.; Pawlik, K.M.; Townes, T.M.; Chen, Y.; Mulloy, J.C.; et al. The AS-RBM15 lncRNA enhances RBM15 protein translation during megakaryocyte differentiation. EMBO Rep. 2016, 17, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Scaria, V.; Hariharan, M.; Maiti, S.; Pillai, B.; Brahmachari, S.K. Host-virus interaction: A new role for microRNAs. Retrovirology 2006, 3, 68. [Google Scholar] [CrossRef] [PubMed]
- Rederstorff, M.; Hüttenhofer, A. Small non-coding RNAs in disease development and host-pathogen interactions. Curr. Opin. Mol. Ther. 2010, 12, 684–694. [Google Scholar] [PubMed]
- Das, K.; Garnica, O.; Dhandayuthapani, S. Modulation of Host miRNAs by Intracellular Bacterial Pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 79. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Kim, T.S.; Basu, J.; Jo, E.-K. MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell. Microbiol. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Zur Bruegge, J.; Einspanier, R.; Sharbati, S. A Long Journey Ahead: Long Non-coding RNAs in Bacterial Infections. Front. Cell. Infect. Microbiol. 2017, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- IIott, N.E.; Heward, J.A.; Roux, B.; Tsitsiou, E.; Fenwick, P.S.; Lenzi, L.; Goodhead, I.; Hertz-Fowler, C.; Heger, A.; Hall, N.; et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2014, 5, 3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leong, H.S.; Dawson, K.; Wirth, C.; Li, Y.; Connolly, Y.; Smith, D.L.; Wilkinson, C.R.M.; Miller, C.J. A global non-coding RNA system modulates fission yeast protein levels in response to stress. Nat. Commun. 2014, 5, 3947. [Google Scholar] [CrossRef] [PubMed]
- Koirala, P.; Huang, J.; Ho, T.-T.; Wu, F.; Ding, X.; Mo, Y.-Y. LncRNA AK023948 is a positive regulator of AKT. Nat. Commun. 2017, 8, 14422. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xue, W.-J.; Feng, Y.; Mao, Q.-S. Long non-coding RNA CASC2 suppresses the proliferation of gastric cancer cells by regulating the MAPK signaling pathway. Am. J. Transl. Res. 2016, 8, 3522–3529. [Google Scholar] [PubMed]
- Li, R.; Zhang, L.; Jia, L.; Duan, Y.; Li, Y.; Bao, L.; Sha, N. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS ONE 2014, 9, e100893. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Vrati, S. The Malat1 long non-coding RNA is upregulated by signalling through the PERK axis of unfolded protein response during flavivirus infection. Sci. Rep. 2015, 5, 17794. [Google Scholar] [CrossRef] [PubMed]

| Method | System | Reference |
|---|---|---|
| Ribosome profiling | macrophages infected with the intracellular bacterial pathogen Legionella pneumophila | [24] |
| Ribosome profiling | macrophages treated with LPS | [25] |
| Sucrose gradient ultracentrifugation followed by microarray analysis | SH-SY5Y cells treated with lytic and sub-lytic doses of α-haemolysin | [14] |
| Pulsed SILAC proteomics | dendritic cells treated with LPS | [26] |
| Sucrose gradient ultracentrifugation followed by PCR array analysis | RAW 264.7 murine macrophages treated with ribotoxic mycotoxin DON | [27] |
| Sucrose gradient ultracentrifugation followed by microarray analysis | human monocyte-derived dendritic cells treated with LPS | [28] |
| Sucrose gradient ultracentrifugation followed by microarray analysis | macrophage-like J774.1 cells treated with LPS | [29] |
| Genomic Position | Mechanism or Function | ||||
|---|---|---|---|---|---|
| Name | Description | Reference | Name | Description | Reference |
| Intergenic lncRNAs (lincRNAs) | do not overlap with any part of a protein coding gene and are at least 1 kb distant from it | [33] | Competing endogenous RNAs (ceRNAs) | also called miRNA “sponges”, which participate in a microRNA-dependent crosstalk. These lncRNAs share miRNA response elements (MREs) with some mRNAs, thereby sequestering miRNAs | [34] |
| Trans-Natural Antisense Transcripts (NATs) | antisense lncRNAs acting on mRNAs and complementary to transcripts from remote loci. | [35] | Protein “sponges” | bind regulatory proteins, disabling them from interacting with their potential targets | [36] |
| Cis-Natural Antisense Transcripts (NATs) | antisense lncRNAs acting on mRNAs. These lncRNAs are transcribed from the same genomic region as their complementary sense transcript | [35] | Scaffolding lncRNAs | act as a scaffold for multiple chromatin remodelling complexes | [37] |
| Sense-overlapping or transcribed pseudogene lncRNAs | are considered transcript variants of protein coding mRNAs, and overlap with a protein coding gene on the same DNA strand | [38] | SINEUPs | antisense lncRNAs that stimulate cap-independent translation of target sense mRNAs through the activity of an embedded repetitive element | [39,40] |
| Intronic lncRNAs | located in the introns of protein coding genes without overlapping with their exons | [41] | Stress-induced lncRNAs (silncRNAs) | Induced upon cell stress, permit a faster recovery of the cell cycle delay caused by stress | [42] |
| Modulators of Post Translational Modifications | Act on post-translational modifications of proteins, such as ubiquitination and phosphorylation | [43] | |||
| Features | Reference |
|---|---|
| Lack of a single long open reading frame (ORF) > 300 nt | [44,45] |
| Low expression levels, compared to mRNAs | [46,47] |
| Longer but fewer exons than protein-coding genes, with a bias toward two-exons transcripts | [48] |
| Exons with a significantly lower GC content, compared to protein-coding RNAs | [44] |
| Paucity or absence of introns | [44] |
| Enrichments in nucleus | [49] |
| High degree of tissue specificity | [46,48] |
| Co-expression with neighboring genes | [46] |
| Low evolutionary conservation of primary sequence | [50] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knap, P.; Tebaldi, T.; Di Leva, F.; Biagioli, M.; Dalla Serra, M.; Viero, G. The Unexpected Tuners: Are LncRNAs Regulating Host Translation during Infections? Toxins 2017, 9, 357. https://doi.org/10.3390/toxins9110357
Knap P, Tebaldi T, Di Leva F, Biagioli M, Dalla Serra M, Viero G. The Unexpected Tuners: Are LncRNAs Regulating Host Translation during Infections? Toxins. 2017; 9(11):357. https://doi.org/10.3390/toxins9110357
Chicago/Turabian StyleKnap, Primoz, Toma Tebaldi, Francesca Di Leva, Marta Biagioli, Mauro Dalla Serra, and Gabriella Viero. 2017. "The Unexpected Tuners: Are LncRNAs Regulating Host Translation during Infections?" Toxins 9, no. 11: 357. https://doi.org/10.3390/toxins9110357
APA StyleKnap, P., Tebaldi, T., Di Leva, F., Biagioli, M., Dalla Serra, M., & Viero, G. (2017). The Unexpected Tuners: Are LncRNAs Regulating Host Translation during Infections? Toxins, 9(11), 357. https://doi.org/10.3390/toxins9110357