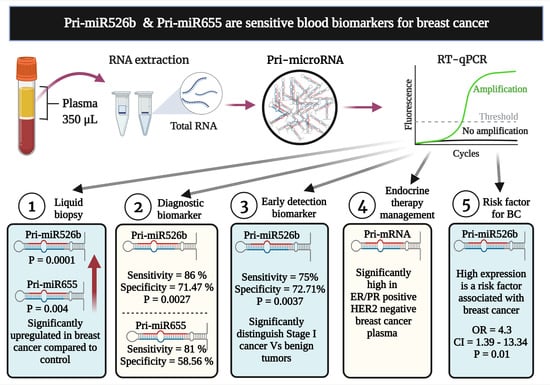
Pri-miR526b and Pri-miR655 Are Potential Blood Biomarkers for Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Patient Demography, and Study Subjects
2.2. Clinical Sample Preparation and Storage
2.3. RNA Extraction and cDNA Synthesis
2.4. Quantitative Real-Time PCR (qRT-PCR)
2.5. Receiver Operating Characteristic (ROC) Curve
2.6. Statistical Analysis
3. Results
3.1. Clinicopathological Characteristics of Benign and Breast Cancer Patients
3.2. Levels of miRNA and Pri-miRNA Expressions Are Comparable in Breast Cancer
3.3. Pri-miR526b and Pri-miR655 Expression in Various Tumor Stages of Plasma Samples
3.4. Diagnostic Performance of Plasma Pri-mir526b and Pri-mir655 in Early Detection of BC
3.5. High Pri-miRNA Expression in Stratified Tumor Samples
3.6. Pri-miRNA as a Risk Factor for Breast Cancer
3.7. Validation of Pri-miRNA Expression in OICR Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fredholm, H.; Eaker, S.; Frisell, J.; Holmberg, L.; Fredriksson, I.; Lindman, H. Breast Cancer in Young Women: Poor Survival Despite Intensive Treatment. PLoS ONE 2009, 4, e7695. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Islami, F.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer in Women: Burden and Trends. Cancer Epidemiol. Biomark. Prev. 2017, 26, 444–457. [Google Scholar] [CrossRef] [Green Version]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global Cancer Statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, G.P.; Major, D.; Chu, C.; Stankiewicz, A.; Harrison, M.L.; Pogany, L.; Mai, V.M.; Onysko, J. A Review of Screening Mammography Participation and Utilization in Canada. Chronic Dis. Inj. Can. 2011, 31, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, L. Early Diagnosis of Breast Cancer. Sensors 2017, 17, 1572. [Google Scholar] [CrossRef] [PubMed]
- Marić, P.; Ozretić, P.; Levanat, S.; Oresković, S.; Antunac, K.; Beketić-Oresković, L. Tumor Markers in Breast Cancer--Evaluation of their Clinical Usefulness. Coll. Antropol. 2011, 35, 241–247. [Google Scholar]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Macfarlane, L.; Murphy, P.R. MicroRNA: Biogenesis, Function, and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [Green Version]
- Wagner, M.; Wiig, H. Tumor Interstitial Fluid Formation, Characterization, and Clinical Implications. Front Oncol. 2015, 5, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Yuan, X.; Shi, H.; Wu, L.; Qian, H.; Xu, W. Exosomes in Cancer: Small Particle, Big Player. J. Hematol. Oncol. 2015, 8, 83. [Google Scholar] [CrossRef] [Green Version]
- Pigati, L.; Yaddanapudi, S.C.S.; Iyengar, R.; Kim, D.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective Release of microRNA Species from Normal and Malignant Mammary Epithelial Cells. PLoS ONE 2010, 5, e13515. [Google Scholar] [CrossRef] [Green Version]
- Adam-Artigues, A.; Garrido-Cano, I.; Simón, S.; Ortega, B.; Moragón, S.; Lameirinhas, A.; Constâncio, V.; Salta, S.; Burgués, O.; Bermejo, B.; et al. Circulating miR-30b-5p Levels in Plasma as a Novel Potential Biomarker for Early Detection of Breast Cancer. ESMO Open 2021, 6, 100039. [Google Scholar] [CrossRef] [PubMed]
- Powrózek, T.; Kuźnar-Kamińska, B.; Dziedzic, M.; Mlak, R.; Batura-Gabryel, H.; Sagan, D.; Krawczyk, P.; Milanowski, J.; Małecka-Massalska, T. The Diagnostic Role of Plasma Circulating Precursors of miRNA-944 and miRNA-3662 for Non-Small Cell Lung Cancer Detection. Pathol. Res. Pract. 2017, 213, 1384–1387. [Google Scholar] [CrossRef]
- Majumder, M.; Xin, X.; Liu, L.; Tutunea-Fatan, E.; Rodriguez-Torres, M.; Vincent, K.; Postovit, L.; Hess, D.; Lala, P.K. COX-2 Induces Breast Cancer Stem Cells Via EP4/PI3K/AKT/NOTCH/WNT Axis. Stem Cells 2016, 34, 2290–2305. [Google Scholar] [CrossRef] [Green Version]
- Majumder, M.; Nandi, P.; Omar, A.; Ugwuagbo, K.C.; Lala, P.K. EP4 as a Therapeutic Target for Aggressive Human Breast Cancer. Int. J. Mol. Sci. 2018, 19, 1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, M.; Landman, E.; Liu, L.; Hess, D.; Lala, P.K. COX-2 Elevates Oncogenic miR-526b in Breast Cancer by EP4 Activation. Mol. Cancer Res. 2015, 13, 1022–1033. [Google Scholar] [CrossRef] [Green Version]
- Majumder, M.; Dunn, L.; Liu, L.; Hasan, A.; Vincent, K.; Brackstone, M.; Hess, D.; Lala, P.K. COX-2 Induces Oncogenic MicroRNA miR655 in Human Breast Cancer. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sourvinou, I.S.; Markou, A.; Lianidou, E.S. Quantification of Circulating miRNAs in Plasma: Effect of Preanalytical and Analytical Parameters on their Isolation and Stability. J. Mol. Diagn. 2013, 15, 827–834. [Google Scholar] [CrossRef]
- Shin, B.; Feser, R.; Nault, B.; Hunter, S.; Maiti, S.; Ugwuagbo, K.C.; Majumder, M. miR526b and miR655 Induce Oxidative Stress in Breast Cancer. Int. J. Mol. Sci 2019, 20, 4039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, S.; Nault, B.; Ugwuagbo, K.C.; Maiti, S.; Majumder, M. Mir526b and Mir655 Promote Tumour Associated Angiogenesis and Lymphangiogenesis in Breast Cancer. Cancers 2019, 11, 938. [Google Scholar] [CrossRef] [Green Version]
- Gervin, E.; Shin, B.; Opperman, R.; Cullen, M.; Feser, R.; Maiti, S.; Majumder, M. Chemically Induced Hypoxia Enhances miRNA Functions in Breast Cancer. Cancers 2020, 12, 2008. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Swets, J.A. ROC Analysis Applied to the Evaluation of Medical Imaging Techniques. Investig. Radiol. 1979, 14, 109–121. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The Meaning and use of the Area Under a Receiver Operating Characteristic (ROC) Curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Winget, M.; Yuan, Y. The Impact of False Positive Breast Cancer Screening Mammograms on Screening Retention: A Retrospective Population Cohort Study in Alberta, Canada. Can. J. Public Health 2018, 108, e539–e545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byers, T.; Wender, R.C.; Jemal, A.; Baskies, A.M.; Ward, E.E.; Brawley, O.W. The American Cancer Society Challenge Goal to Reduce US Cancer Mortality by 50% between 1990 and 2015: Results and Reflections. CA Cancer J. Clin. 2016, 66, 359–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of mRNAs and microRNAs is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of microRNAs Detectable in Serum and Saliva is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [Green Version]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as Novel Minimally Invasive Biomarkers for Breast Cancer. Ann. Surg. 2010, 251, 499–505. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene Expression Patterns of Breast Carcinomas Distinguish Tumor Subclasses with Clinical Implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [Green Version]
- Hosseini Mojahed, F.; Aalami, A.H.; Pouresmaeil, V.; Amirabadi, A.; Qasemi Rad, M.; Sahebkar, A. Clinical Evaluation of the Diagnostic Role of MicroRNA-155 in Breast Cancer. Int. J. Genom. 2020, 2020, 9514831. [Google Scholar] [CrossRef]
- McVeigh, T.P.; Mulligan, R.J.; McVeigh, U.M.; Owens, P.W.; Miller, N.; Bell, M.; Sebag, F.; Guerin, C.; Quill, D.S.; Weidhaas, J.B.; et al. Investigating the Association of rs2910164 with Cancer Predisposition in an Irish Cohort. Endocr. Connect. 2017, 6, 614–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Liang, J.; Wang, Z.; Tian, T.; Zhou, X.; Chen, J.; Miao, R.; Wang, Y.; Wang, X.; Shen, H. Common Genetic Variants in Pre-microRNAs were Associated with Increased Risk of Breast Cancer in Chinese Women. Hum. Mutat. 2009, 30, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Caselli, G.; Bonazzi, A.; Lanza, M.; Ferrari, F.; Maggioni, D.; Ferioli, C.; Giambelli, R.; Comi, E.; Zerbi, S.; Perrella, M.; et al. Pharmacological Characterisation of CR6086, a Potent Prostaglandin E2 Receptor 4 Antagonist, as a New Potential Disease-Modifying Anti-Rheumatic Drug. Arthritis Res. Ther. 2018, 20, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Sample No. | Pathology | Histopathological Details | Age | Diagnosed with Cancer after Specimen Collection |
|---|---|---|---|---|
| C1 | Benign | Atypical ductal hyperplasia within a complex, sclerosed papilloma; fibroadenoma | 59 | No |
| C2 | Benign | Fibrosis intermixed with foamy macrophages and numerous stromal microcalcifications, suggestive of healing fat necrosis | 90 | No |
| C3 | Benign | Benign phyllodes tumor | 51 | No |
| C4 | Benign | The lesion is a benign fibroadenoma, but the patient has a recent history of breast cancer within the past 6 months; undergoing adjuvant treatment at the time of this biopsy | 55 | No |
| C5 | Benign | Fibroadenoma with focal with florid ductal hyperplasia | 52 | No |
| C6 | Benign | Focal cyst rupture, with associated chronic inflammation, fibrosis, collections of foamy histiocytes, and multinucleated giant cell reaction | 59 | Yes, in 2018 |
| C7 | Benign | Papillary lesion without atypia | 82 | No |
| C8 | Benign | Nonproliferative change | 50 | No |
| C9 | Benign | Fibrocystic and fibro adenomatoid changes, focal epithelial hyperplasia | 51 | No |
| C10 | Benign | Fragments of reactive lymph node tissue, unremarkable breast parenchyma | 61 | No |
| C11 | Benign | Biopsy specimen: nonproliferative (fibrocystic) change (fibro adenomatoid hyperplasia, cysts, fibrosis); lumpectomy specimen: proliferative breast disease without atypia (moderate ductal epithelial hyperplasia, columnar cell type); regional dense stromal fibrosis | 44 | No |
| C12 | Benign | Papilloma | 60 | No |
| C13 | Benign | Benign fibro glandular tissue with focus of inflammation suggestive of resolving fat necrosis | 64 | No |
| C14 | Benign | Focal abscess with mixed acute and chronic inflammation, histiocytes, and fibrosis | 36 | No |
| C15 | Benign | Atypical ductal hyperplasia (ADH) involving a complex intraductal papilloma | 46 | Yes, in 2017 |
| C16 | Benign | Sclerosed complex papillary lesion | 57 | No |
| C17 | Benign | Fibroadenoma | 46 | No |
| C18 | Benign | Fat necrosis with calcifications, florid ductal epithelial hyperplasia | 48 | No |
| C19 | Benign | Smooth muscle tumor | 63 | No |
| C20 | Benign | Complex papillary lesion without atypia | 59 | No |
| Characteristics | Control n = 20 | Breast Cancer n = 90 (100%) | |
|---|---|---|---|
| Sex | Male | 0 | 0 |
| Female | 20 | 90 | |
| Age (years) | Mean ± SD (range) | 66 ± 11 (52–87) | 64 ± 12 (42–91) |
| Tumor Receptor Status | ER+ve (%) | 73 (81.11) | |
| ER−ve (%) | 17 (18.89) | ||
| PR+ve (%) | 68 (75.56) | ||
| PR−ve (%) | 22 (24.44) | ||
| HER2+ve (%) | 19 (21.11) | ||
| HER2−ve (%) | 71 (78.89) | ||
| ER/PR/HER2−ve (%) | 8 (8.89) | ||
| TNM (Tumor) Staging | 0 | 1 (1.11) | |
| I | 43 (47.78) | ||
| II | 36 (40.00) | ||
| III | 8 (8.89) | ||
| IV | 2 (2.22) | ||
| Mean Ct Cut-Off | Sensitivity (%) | Specificity (%) | PPV (95% CI) | NPV (95% CI) | Accuracy (95% CI) | |
|---|---|---|---|---|---|---|
| Pri-miR526b | 6.73 | 86.02 | 41.18 | 88.89 (80.51–94.54) | 35.00 (15.39–59.22) | 80.70 (71.30–87.02) |
| Pri-miR655 | 3.63 | 80.85 | 12.5 | 84.44 (75.28–91.23) | 10.00 (1.24–31.70) | 71.82 (62.44–79.98) |
| Unstratified Tumor vs. Control | Stratified Tumor (Stage I) vs. Control | |||
|---|---|---|---|---|
| Pri-miR526b | Pri-miR655 | Pri-miR526b | Pri-miR655 | |
| AUC (95% CI) | 71.47 (59.80–83.14) | 58.56 (46.86–70.25) | 72.73 (59.84–85.62) | 59.20 (45.40–73.01) |
| p-value | 0.0027 | 0.2327 | 0.0037 | 0.2408 |
| Sensitivity (95% CI) | 86.02 (77.28–92.34) | 80.85 (71.44–88.24) | 75.00 (61.05–85.97) | 67.27 (53.29–79.32) |
| Specificity (95% CI) | 41.18 (18.44–67.08) | 12.50 (1.55–38.35) | 58.33 (27.67–84.83) | 22.22 (2.815–60.10) |
| Tumor Status | No. of Samples n (%) | High Pri-miR526b n (%) | Z-Score | p-Value | High Pri-miR655 n (%) | Z-Score | p-Value |
|---|---|---|---|---|---|---|---|
| ER+ve | 73 (81.1) | 60 (82.1) | 2.0872 | 0.0366 | 58 (79.4) | 2.7201 | 0.0065 |
| ER−ve | 17 (18.9) | 10 (58.8) | 8 (47.0) | ||||
| PR+ve | 68 (75.6) | 55 (80.9) | 1.2455 | 0.2113 | 53 (77.9) | 1.7379 | 0.0819 |
| PR−ve | 22 (24.4) | 15 (68.2) | 13 (59.1) | ||||
| HER2+ve | 19 (21.1) | 10 (52.6) | −2.7888 | 0.0053 | 10 (52.6) | −2.1465 | 0.0316 |
| HER2−ve | 71 (78.9) | 59 (83.1) | 55 (77.5) |
| Ct Cutoff | BC High Pri-miRNA | Benign High Pri-miRNA | Odds Ratio | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| Pri-miR526b | 6.73 | 80 | 13 | 4.308 | 1.391–13.34 | 0.0142 |
| Pri-miR655 | 3.63 | 76 | 18 | 0.6032 | 0.126–2.895 | 0.7316 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumder, M.; Ugwuagbo, K.C.; Maiti, S.; Lala, P.K.; Brackstone, M. Pri-miR526b and Pri-miR655 Are Potential Blood Biomarkers for Breast Cancer. Cancers 2021, 13, 3838. https://doi.org/10.3390/cancers13153838
Majumder M, Ugwuagbo KC, Maiti S, Lala PK, Brackstone M. Pri-miR526b and Pri-miR655 Are Potential Blood Biomarkers for Breast Cancer. Cancers. 2021; 13(15):3838. https://doi.org/10.3390/cancers13153838
Chicago/Turabian StyleMajumder, Mousumi, Kingsley Chukwunonso Ugwuagbo, Sujit Maiti, Peeyush K Lala, and Muriel Brackstone. 2021. "Pri-miR526b and Pri-miR655 Are Potential Blood Biomarkers for Breast Cancer" Cancers 13, no. 15: 3838. https://doi.org/10.3390/cancers13153838
APA StyleMajumder, M., Ugwuagbo, K. C., Maiti, S., Lala, P. K., & Brackstone, M. (2021). Pri-miR526b and Pri-miR655 Are Potential Blood Biomarkers for Breast Cancer. Cancers, 13(15), 3838. https://doi.org/10.3390/cancers13153838