Nanolayers of Poly(N,N′-Dimethylaminoethyl Methacrylate) with a Star Topology and Their Antibacterial Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of the N,N′-Dimethylaminoethyl Methacrylate Star and Linear Polymer
2.3. Preparation of Benzophenone Modified Wafers
2.4. Formation of Polymer Layers on the Glass and Silicon Wafers with UV Irradiation
2.5. Quaternization of the Linear and Star PDMAEMA
2.6. Evaluation of Antibacterial Properties
2.7. Characterization Methods
3. Results and Discussion
3.1. Formation of the Layers Made of the Star and Linear Polymers on a Solid Surface
3.2. Physicochemical Characterization of the Obtained PDMAEMA Layers
3.3. Quaternization of the PDMAEMA Nanolayers
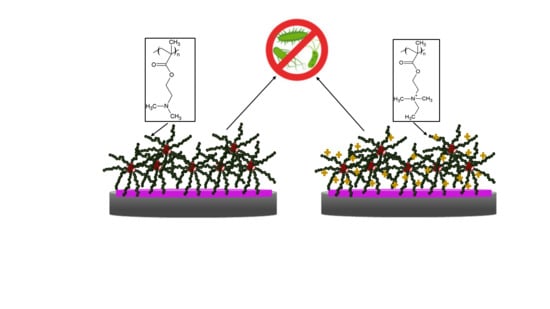
3.4. Antibacterial Activity of PDMAEMA and QPDMAEMA Nanolayers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hasan, J.; Crawford, R.J.; Ivanova, E.P. Antibacterial surfaces: The quest for a new generation of biomaterials. Trends Biotechnol. 2013, 31, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, L.M.; Kleshcheva, N.A.; Moroz, A.F.; Didenko, L.V. Secondary and Tertiary Polydiallylammonium Salts: Novel Polymers with High Antimicrobial Activity. Biomacromolecules 2009, 10, 2976–2986. [Google Scholar] [CrossRef] [PubMed]
- Vigliotta, G.; Mella, M.; Rega, D.; Izzo, L. Modulating Antimicrobial Activity by Synthesis: Dendritic Copolymers Based on Nonquaternized 2-(Dimethylamino)ethyl Methacrylate by Cu-Mediated ATRP. Biomacromolecules 2012, 13, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Siedenbiedel, F.; Fuchs, A.; Moll, T.; Weide, M.; Breves, R.; Tiller, J.C. Star-Shaped Poly(styrene)-block-Poly(4-vinyl-N-methylpyridiniumiodide) for Semipermanent Antimicrobial Coatings. Macromol. Biosci. 2013, 13, 1447–1455. [Google Scholar] [CrossRef] [PubMed]
- Borisova, D.; Haladjova, E.; Kyulavska, M.; Petrov, P.; Pispas, S.; Stoitsova, S.; Paunova-Krasteva, T. Application of cationic polymer micelles for the dispersal of bacterial biofilms. Eng. Life Sci. 2018, 18, 943–948. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, I.; Ghosh, A.; Bhadury, P.; De, P. Leucine-Based Polymer Architecture-Induced Antimicrobial Properties and Bacterial Cell Morphology Switching. ACS Omega 2018, 3, 769–780. [Google Scholar] [CrossRef]
- Kenawy, E.-R.; Bowlin, G.L.; Mansfield, K.; Layman, J.; Simpson, D.G.; Sanders, E.H.; Wnek, G.E. Release of tetracycline hydrochloride from electrospun poly(ethylene-co-vinylacetate), poly(lactic acid), and a blend. J. Control. Release 2002, 81, 57–64. [Google Scholar] [CrossRef]
- Buschle-Diller, G.; Cooper, J.; Xie, Z.; Wu, Y.; Waldrup, J.; Ren, X. Release of antibiotics from electrospun bicomponent fibers. Cellulose 2007, 14, 553–562. [Google Scholar] [CrossRef]
- Kalyon, B.D.; Olgun, U. Antibacterial efficacy of triclosan-incorporated polymers. Am. J. Infect. Control 2001, 29, 124–125. [Google Scholar] [CrossRef]
- Tallury, P.; Airrabeelli, R.; Li, J.; Paquette, D.; Kalachandra, S. Release of antimicrobial and antiviral drugs from methacrylate copolymer system: Effect of copolymer molecular weight and drug loading on drug release. Dent. Mater. 2008, 24, 274–280. [Google Scholar] [CrossRef]
- Son, Y.-A.; Sun, G. Durable antimicrobial nylon 66 fabrics: Ionic interactions with quaternary ammonium salts. J. Appl. Polym. Sci. 2003, 90, 2194–2199. [Google Scholar] [CrossRef]
- Ramstedt, M.; Cheng, N.; Azzaroni, O.; Mossialos, D.; Mathieu, H.J.; Huck, W.T.S. Synthesis and Characterization of Poly(3-Sulfopropylmethacrylate) Brushes for Potential Antibacterial Applications. Langmuir 2007, 23, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H.; Odermatt, E.K.; Berndt, I.; Tiller, J.C. Long-term active antimicrobial coatings for surgical sutures based on silver nanoparticles and hyperbranched polylysine. J. Biomater. Sci. Polym. Ed. 2013, 24, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Wang, C.; Wang, Y.; Xu, W.; Hu, J.; Cheng, Y. A Nanocomposite Hydrogel with Potent and Broad-Spectrum Antibacterial Activity. ACS Appl. Mater. Interfaces 2018, 10, 15163–15173. [Google Scholar] [CrossRef]
- Rasool, K.; Nasrallah, G.K.; Younes, N.; Pandey, R.P.; Abdul Rasheed, P.; Mahmoud, K.A. “Green” ZnO-Interlinked Chitosan Nanoparticles for the Efficient Inhibition of Sulfate-Reducing Bacteria in Inject Seawater. ACS Sustain. Chem. Eng. 2018, 6, 3896–3906. [Google Scholar] [CrossRef]
- Xiu, K.; Wen, J.; Liu, J.; He, C.; Sun, Y. Controlling the Structure and Antimicrobial Function of N-Halamine-Based Polyurethane Semi-interpenetrating Polymer Networks. Ind. Eng. Chem. Res. 2017, 56, 12032–12037. [Google Scholar] [CrossRef]
- Hong, W.; Zhao, Y.; Guo, Y.; Huang, C.; Qiu, P.; Zhu, J.; Chu, C.; Shi, H.; Liu, M. PEGylated Self-Assembled Nano-Bacitracin A: Probing the Antibacterial Mechanism and Real-Time Tracing of Target Delivery in Vivo. ACS Appl. Mater. Interfaces 2018, 10, 10688–10705. [Google Scholar] [CrossRef]
- Mauro, N.; Schillaci, D.; Varvarà, P.; Cusimano, M.G.; Geraci, D.M.; Giuffrè, M.; Cavallaro, G.; Maida, C.M.; Giammona, G. Branched High Molecular Weight Glycopolypeptide With Broad-Spectrum Antimicrobial Activity for the Treatment of Biofilm Related Infections. ACS Appl. Mater. Interfaces 2018, 10, 318–331. [Google Scholar] [CrossRef]
- Liu, M.; Li, J.; Li, B. Mannose-Modificated Polyethylenimine: A Specific and Effective Antibacterial Agent against Escherichia coli. Langmuir 2018, 34, 1574–1580. [Google Scholar] [CrossRef]
- Wang, X.; Yan, S.; Song, L.; Shi, H.; Yang, H.; Luan, S.; Huang, Y.; Yin, J.; Khan, A.F.; Zhao, J. Temperature-Responsive Hierarchical Polymer Brushes Switching from Bactericidal to Cell Repellency. ACS Appl. Mater. Interfaces 2017, 9, 40930–40939. [Google Scholar] [CrossRef]
- Waschinski, C.J.; Herdes, V.; Schueler, F.; Tiller, J.C. Influence of Satellite Groups on Telechelic Antimicrobial Functions of Polyoxazolines. Macromol. Biosci. 2005, 5, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Waschinski, C.J.; Barnert, S.; Theobald, A.; Schubert, R.; Kleinschmidt, F.; Hoffmann, A.; Saalwächter, K.; Tiller, J.C. Insights in the Antibacterial Action of Poly(methyloxazoline)s with a Biocidal End Group and Varying Satellite Groups. Biomacromolecules 2008, 9, 1764–1771. [Google Scholar] [CrossRef] [PubMed]
- Fik, C.P.; Krumm, C.; Muennig, C.; Baur, T.I.; Salz, U.; Bock, T.; Tiller, J.C. Impact of Functional Satellite Groups on the Antimicrobial Activity and Hemocompatibility of Telechelic Poly(2-methyloxazoline)s. Biomacromolecules 2012, 13, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Konai, M.M.; Bhattacharjee, B.; Ghosh, S.; Haldar, J. Recent Progress in Polymer Research to Tackle Infections and Antimicrobial Resistance. Biomacromolecules 2018, 19, 1888–1917. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Tang, Z.; Yu, Q.; Chen, H. Smart Antibacterial Surfaces with Switchable Bacteria-Killing and Bacteria-Releasing Capabilities. ACS Appl. Mater. Interfaces 2017, 9, 37511–37523. [Google Scholar] [CrossRef]
- Krishnamoorthy, M.; Hakobyan, S.; Ramstedt, M.; Gautrot, J.E. Surface-Initiated Polymer Brushes in the Biomedical Field: Applications in Membrane Science, Biosensing, Cell Culture, Regenerative Medicine and Antibacterial Coatings. Chem. Rev. 2014, 114, 10976–11026. [Google Scholar] [CrossRef]
- Wu, W.; Wang, W.; Li, J. Star polymers: Advances in biomedical applications. Prog. Polym. Sci. 2015, 46, 55–85. [Google Scholar] [CrossRef]
- Jiao, Y.; Niu, L.-N.; Ma, S.; Li, J.; Tay, F.R.; Chen, J.-H. Quaternary ammonium-based biomedical materials: State-of-the-art, toxicological aspects and antimicrobial resistance. Prog. Polym. Sci. 2017, 71, 53–90. [Google Scholar] [CrossRef]
- Muñoz-Bonilla, A.; Fernández-García, M. Polymeric materials with antimicrobial activity. Prog. Polym. Sci. 2012, 37, 281–339. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, M.A.; De Melo Carrasco, L.D. Cationic Antimicrobial Polymers and Their Assemblies. Int. J. Mol. Sci. 2013, 14, 9906–9946. [Google Scholar] [CrossRef] [Green Version]
- Kügler, R.; Bouloussa, O.; Rondelez, F. Evidence of a charge-density threshold for optimum efficiency of biocidal cationic surfaces. Microbiology 2005, 151, 1341–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Qiu, S.; Lewis, K.; Klibanov, A.M. Bactericidal Properties of Flat Surfaces and Nanoparticles Derivatized with Alkylated Polyethylenimines. Biotechnol. Prog. 2002, 18, 1082–1086. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Qiu, S.; Lewis, K.; Klibanov, A.M. Mechanism of bactericidal and fungicidal activities of textiles covalently modified with alkylated polyethylenimine. Biotechnol. Bioeng. 2003, 83, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, H.; Tian, Z.; Sen, A.; Allcock, H.R. Preparation of quaternized organic–inorganic hybrid brush polyphosphazene-co-poly[2 -(dimethylamino)ethyl methacrylate] electrospun fibers and their antibacterial properties. Polym. Chem. 2012, 3, 2082–2091. [Google Scholar] [CrossRef]
- Mendrek, B.; Sieroń, Ł.; Żymełka-Miara, I.; Binkiewicz, P.; Libera, M.; Smet, M.; Trzebicka, B.; Sieroń, A.L.; Kowalczuk, A.; Dworak, A. Nonviral Plasmid DNA Carriers Based on N,N′-Dimethylaminoethyl Methacrylate and Di(ethylene glycol) Methyl Ether Methacrylate Star Copolymers. Biomacromolecules 2015, 16, 3275–3285. [Google Scholar] [CrossRef]
- Zhang, Y.; He, X.; Ding, M.; He, W.; Li, J.; Li, J.; Tan, H. Antibacterial and Biocompatible Cross-Linked Waterborne Polyurethanes Containing Gemini Quaternary Ammonium Salts. Biomacromolecules 2018, 19, 279–287. [Google Scholar] [CrossRef]
- Chen, S.; Chen, Q.; Li, Q.; An, J.; Sun, P.; Ma, J.; Gao, H. Biodegradable Synthetic Antimicrobial with Aggregation-Induced Emissive Luminogens for Temporal Antibacterial Activity and Facile Bacteria Detection. Chem. Mater. 2018, 30, 1782–1790. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, G.; Ge, Z.; Liu, S. Stimuli-responsive tertiary amine methacrylate-based block copolymers: Synthesis, supramolecular self-assembly and functional applications. Prog. Polym. Sci. 2014, 39, 1096–1143. [Google Scholar] [CrossRef]
- Huang, J.; Koepsel, R.R.; Murata, H.; Wu, W.; Lee, S.B.; Kowalewski, T.; Russell, A.J.; Matyjaszewski, K. Nonleaching Antibacterial Glass Surfaces via “Grafting Onto”: The Effect of the Number of Quaternary Ammonium Groups on Biocidal Activity. Langmuir 2008, 24, 6785–6795. [Google Scholar] [CrossRef]
- Dong, Z.; Wei, H.; Mao, J.; Wang, D.; Yang, M.; Bo, S.; Ji, X. Synthesis and responsive behavior of poly(N,N-dimethylaminoethyl methacrylate) brushes grafted on silica nanoparticles and their quaternized derivatives. Polymer 2012, 53, 2074–2084. [Google Scholar] [CrossRef]
- Mazloomi-Rezvani, M.; Salami-Kalajahi, M.; Roghani-Mamaqani, H. “Grafting to” approach for surface modification of AuNPs with RAFT-mediated synthesized smart polymers: Stimuli-responsive behaviors of hybrid nanoparticles. J. Phys. Chem. Solids 2018, 123, 183–190. [Google Scholar] [CrossRef]
- Lee, S.B.; Koepsel, R.R.; Morley, S.W.; Matyjaszewski, K.; Sun, Y.; Russell, A.J. Permanent, Nonleaching Antibacterial Surfaces. 1. Synthesis by Atom Transfer Radical Polymerization. Biomacromolecules 2004, 5, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Knapp, J.S.; Guthrie, J.T.; Perrier, S. Antibacterial Cellulose Fiber via RAFT Surface Graft Polymerization. Biomacromolecules 2008, 9, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhu, X.; Shi, Z.L.; Neoh, K.G.; Kang, E.T. Polymer Microspheres with Permanent Antibacterial Surface from Surface-Initiated Atom Transfer Radical Polymerization. Ind. Eng. Chem. Res. 2005, 44, 7098–7104. [Google Scholar] [CrossRef]
- Yuan, S.J.; Pehkonen, S.O.; Ting, Y.P.; Neoh, K.G.; Kang, E.T. Antibacterial Inorganic–Organic Hybrid Coatings on Stainless Steel via Consecutive Surface-Initiated Atom Transfer Radical Polymerization for Biocorrosion Prevention. Langmuir 2010, 26, 6728–6736. [Google Scholar] [CrossRef]
- Murata, H.; Koepsel, R.R.; Matyjaszewski, K.; Russell, A.J. Permanent, non-leaching antibacterial surfaces-2: How high density cationic surfaces kill bacterial cells. Biomaterials 2007, 28, 4870–4879. [Google Scholar] [CrossRef]
- Huang, J.; Murata, H.; Koepsel, R.R.; Russell, A.J.; Matyjaszewski, K. Antibacterial Polypropylene via Surface-Initiated Atom Transfer Radical Polymerization. Biomacromolecules 2007, 8, 1396–1399. [Google Scholar] [CrossRef]
- Li, M.; Liu, X.; Liu, N.; Guo, Z.; Singh, P.K.; Fu, S. Effect of surface wettability on the antibacterial activity of nanocellulose-based material with quaternary ammonium groups. Colloids Surf. A Physicochem. Eng. Asp. 2018, 554, 122–128. [Google Scholar] [CrossRef]
- Weng, Y.; Guo, X.; Zhao, J.; Gregory, R.L.; Xie, D. A PQAS-containing glass-ionomer cement for improved antibacterial function. J. Biomed. Sci. Eng. 2010, 3, 955–962. [Google Scholar] [CrossRef] [Green Version]
- Weng, Y.; Howard, L.; Chong, V.J.; Sun, J.; Gregory, R.L.; Xie, D. A novel furanone-modified antibacterial dental glass ionomer cement. Acta Biomater. 2012, 8, 3153–3160. [Google Scholar] [CrossRef]
- Prucker, O.; Naumann, C.A.; Rühe, J.; Knoll, W.; Frank, C.W. Photochemical Attachment of Polymer Films to Solid Surfaces via Monolayers of Benzophenone Derivatives. J. Am. Chem. Soc. 1999, 121, 8766–8770. [Google Scholar] [CrossRef]
- Mendrek, B.; Sieroń, Ł.; Libera, M.; Smet, M.; Trzebicka, B.; Sieroń, A.L.; Dworak, A.; Kowalczuk, A. Polycationic star polymers with hyperbranched cores for gene delivery. Polymer 2014, 55, 4551–4562. [Google Scholar] [CrossRef]
- Kowalczuk, A.; Vandendriessche, A.; Trzebicka, B.; Mendrek, B.; Szeluga, U.; Cholewiński, G.; Smet, M.; Dworak, A.; Dehaen, W. Core-shell nanoparticles with hyperbranched poly(arylene-oxindole) interiors. J. Polym. Sci. A Polym. Chem. 2009, 47, 1120–1135. [Google Scholar] [CrossRef]
- Zhang, X.; Xia, J.; Matyjaszewski, K. Controlled/“Living” Radical Polymerization of 2-(Dimethylamino)ethyl Methacrylate. Macromolecules 1998, 31, 5167–5169. [Google Scholar] [CrossRef] [PubMed]
- Utrata-Wesołek, A.; Oleszko, N.; Trzebicka, B.; Anioł, J.; Zagdańska, M.; Lesiak, M.; Sieroń, A.; Dworak, A. Modified polyglycidol based nanolayers of switchable philicity and their interactions with skin cells. Eur. Polym. J. 2013, 49, 106–117. [Google Scholar] [CrossRef]
- Oleszko, N.; Wałach, W.; Utrata-Wesołek, A.; Kowalczuk, A.; Trzebicka, B.; Klama-Baryła, A.; Hoff-Lenczewska, D.; Kawecki, M.; Lesiak, M.; Sieroń, A.L.; et al. Controlling the Crystallinity of Thermoresponsive Poly(2-oxazoline)-Based Nanolayers to Cell Adhesion and Detachment. Biomacromolecules 2015, 16, 2805–2813. [Google Scholar] [CrossRef] [PubMed]
- Plamper, F.A.; Schmalz, A.; Penott-Chang, E.; Drechsler, M.; Jusufi, A.; Ballauff, M.; Müller, A.H.E. Synthesis and Characterization of Star-Shaped Poly(N,N-dimethylaminoethyl methacrylate) and Its Quaternized Ammonium Salts. Macromolecules 2007, 40, 5689–5697. [Google Scholar] [CrossRef]
- Nanci, A.; Wuest, J.D.; Peru, L.; Brunet, P.; Sharma, V.; Zalzal, S.; McKee, M.D. Chemical modification of titanium surfaces for covalent attachment of biological molecules. J. Biomed. Mater. Res. 1998, 40, 324–335. [Google Scholar] [CrossRef]
- Mendrek, B.; Żymełka-Miara, I.; Sieroń, Ł.; Fus, A.; Balin, K.; Kubacki, J.; Smet, M.; Trzebicka, B.; Sieroń, A.L.; Kowalczuk, A. Stable star polymer nanolayers and their thermoresponsiveness as a tool for controlled culture and detachment of fibroblast sheets. J. Mater. Chem. B 2018, 6, 641–655. [Google Scholar] [CrossRef]
- Mendrek, B. Behavior of methacrylate star copolymers in solutions. Polimery 2016, 6, 413–419. [Google Scholar] [CrossRef]
- Dong, H.; Huang, J.; Koepsel, R.R.; Ye, P.; Russell, A.J.; Matyjaszewski, K. Recyclable Antibacterial Magnetic Nanoparticles Grafted with Quaternized Poly(2-(dimethylamino)ethyl methacrylate) Brushes. Biomacromolecules 2011, 12, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Tu, Q.; Tian, C.; Ma, T.; Pang, L.; Wang, J. Click synthesis of quaternized poly(dimethylaminoethyl methacrylate) functionalized graphene oxide with improved antibacterial and antifouling ability. Colloids Surf. B Biointerfaces 2016, 141, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Wahlen, L.; Mantei, J.R.; DiOrio, J.P.; Jones, C.M.; Pasmore, M.E. Production and analysis of a Bacillus subtilis biofilm comprised of vegetative cells and spores using a modified colony biofilm model. J. Microbiol. Methods 2018, 148, 181–187. [Google Scholar] [CrossRef] [PubMed]









| PDMAEMA | Mn GPC-MALLS (g/mol) | Mw/Mn |
|---|---|---|
| L1 | 9000 | 1.09 |
| L2 | 13,000 | 1.10 |
| L3 | 16,000 | 1.08 |
| L4 | 40,000 | 1.12 |
| G1 | 320,000 | 2.70 |
| G2 | 400,000 | 2.90 |
| G3 | 560,000 | 2.33 |
| G4 | 1,000,000 | 1.79 |
| Sample | C 1s (%) | N 1s (%) | O 1s (%) | Br 3d (%) | C/O |
|---|---|---|---|---|---|
| SOH | 18.3 | 1.9 | 79.8 | 0.0 | 0.2 |
| SBPH | 28.3 | 0.2 | 71.5 | 0.0 | 0.4 |
| SL3 | 65.9 | 5.0 | 29.0 | 0.1 | 2.3 |
| SG3 | 74.9 | 5.1 | 19.7 | 0.3 | 3.8 |
| Layer | Layer Thickness-AFM (nm) | Layer thickness-Ellipsometry (nm) |
|---|---|---|
| SL1 | 4 | 3 |
| SL2 | 6 | 4 |
| SL3 | 7 | 6 |
| SL4 | 13 | 10 |
| SG1 | 31 | 50 |
| SG2 | 78 | 70 |
| SG3 | 70 | 87 |
| SG4 | 120 | 100 |
| Nanolayers | Concentration of Quaternized Amino Groups on the Surface (mg/mL/cm2) | Quantity of Quaternized Amino Groups on the cm2 Surface |
|---|---|---|
| QSL2 | 0.005 | 2.40 × 1016 |
| QSL4 | 0.031 | 1.49 × 1017 |
| QSG2 | 0.323 | 1.55 × 1018 |
| QSG4 | 0.463 | 2.22 × 1018 |
| Nanolayer | Number of Bacteria (CFU/mL) 5 min | Growth Inhibition (%) 5 min | Number of Bacteria (CFU/mL) 60 min | Growth Inhibition (%) 60 min | Number of Bacteria (CFU/mL) 24 h | Growth Inhibition (%) 24 h |
|---|---|---|---|---|---|---|
| SG4 | 1.1 × 106 | 78% | 1.01 × 106 | 80% | 6.1 × 105 | 89% |
| QSG4 | 3.3 × 105 | 93% | 2.5 × 106 | 49% | 7.2 × 106 | 0% |
| SL4 | 6.2 × 105 | 88% | 6.9 × 106 | 81% | 2.4 × 106 | 62% |
| QSL4 | 1.1 × 106 | 79% | 1.6 × 107 | 0% | 1.4 × 106 | 0% |
| Control | 5 × 106 | - | 5.2 × 106 | - | 6.2 × 106 | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teper, P.; Chojniak-Gronek, J.; Hercog, A.; Oleszko-Torbus, N.; Płaza, G.; Kubacki, J.; Balin, K.; Kowalczuk, A.; Mendrek, B. Nanolayers of Poly(N,N′-Dimethylaminoethyl Methacrylate) with a Star Topology and Their Antibacterial Activity. Polymers 2020, 12, 230. https://doi.org/10.3390/polym12010230
Teper P, Chojniak-Gronek J, Hercog A, Oleszko-Torbus N, Płaza G, Kubacki J, Balin K, Kowalczuk A, Mendrek B. Nanolayers of Poly(N,N′-Dimethylaminoethyl Methacrylate) with a Star Topology and Their Antibacterial Activity. Polymers. 2020; 12(1):230. https://doi.org/10.3390/polym12010230
Chicago/Turabian StyleTeper, Paulina, Joanna Chojniak-Gronek, Anna Hercog, Natalia Oleszko-Torbus, Grażyna Płaza, Jerzy Kubacki, Katarzyna Balin, Agnieszka Kowalczuk, and Barbara Mendrek. 2020. "Nanolayers of Poly(N,N′-Dimethylaminoethyl Methacrylate) with a Star Topology and Their Antibacterial Activity" Polymers 12, no. 1: 230. https://doi.org/10.3390/polym12010230
APA StyleTeper, P., Chojniak-Gronek, J., Hercog, A., Oleszko-Torbus, N., Płaza, G., Kubacki, J., Balin, K., Kowalczuk, A., & Mendrek, B. (2020). Nanolayers of Poly(N,N′-Dimethylaminoethyl Methacrylate) with a Star Topology and Their Antibacterial Activity. Polymers, 12(1), 230. https://doi.org/10.3390/polym12010230