Unlocking the Potential of Molecularly Imprinted Polydopamine in Sensing Applications
Abstract
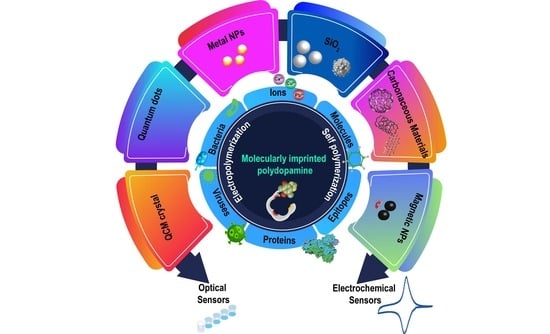
:1. Introduction
2. Core/Substrate Materials Used to Be Modified with Molecularly Imprinted Polydopamine (MIPda)
| Substrate Materials | Configuration | Template | Synthesis Method | Morphology | Role of the Core/Support Materials | Comments |
|---|---|---|---|---|---|---|
| Magnetic Nanoparticles | Fe3O4 NPs [34] | Thionine | Self-polymerization if the presence of saturated O2 solution | uniform spherical particles and smooth surface of Fe3O4-MIPda Fe3O4 NPs: 600 nm Fe3O4-MIPda: 600 nm (thin layer) | Nanozyme Magnetic properties | Fe3O4 NPs were prepared by the solvothermal method |
| Fe3O4@SO2 [35] | ovalbumin | Self-polymerization | spherical and relatively uniform MNPs: 13 nm Fe3O4@SiO2: 73 nm Fe3O4@SiO2@MIPda: NA | Magnetic properties | SiO2 provides hydroxyl groups and protects Fe3O4 from oxidation and acid attack during the removal of templates | |
| Fe3O4@fibrous SiO2 [36] | lysozyme | Self-polymerization | The MIPDA layer evenly covered the fibrous SiO2 surface. The average diameter of the MIP-lysozyme microsphere is 445 nm, indicating an average PDA imprinting layer thickness of 45 nm | Magnetic properties and high surface area (570 m2/g) allowing for fast adsorption | The PDA layer exhibits a photothermal effect and shows the controlled release property triggered by NIR laser | |
| Fe3O4/PMMA [37] | lysozyme | Self-polymerization | Fe3O4: 6 nm Fe3O4/PMMA: 150 nm Fe3O4/PMMA/MIP; 180 nm | Magnetic properties and hydroxyl groups | Fe3O4/PMMA was prepared via miniemulsion and the lysozyme was immobilized on the surface of Fe3O4/PMMA. However, PMMA is not good support for immobilizing lysozyme | |
| Metal/metal oxide | CdSe–CdS–Zn/Ti substrate [39] | L-phenylalanine | Electropolymerization | CdS/CdSe layer: spherical structure; CdS/CdSe–MIPda: spherical smooth ad compact | Photoelectrochemical properties | The preparation of the electrode is complicated |
| glassy carbon electrode modified with AuNPs [40] | Pseudomonas aeruginosa | electropolymerization | uniform MIP | AuNPs were employed as a specialized intermediary and interface to enhance the loading rate of the aptamer sequence | The dual precise molecular recognition characteristics of both MIP and aptamers resulted in exceptional sensing capabilities | |
| AuNP-coated screen-printed carbon electrode [41] | The igE-binding epitope of ovalbumin | electropolymerization | spherical-like cluster | electrocatalytic activity | The limited electroactivity of MIPda was countered by electro-depositing AuNPs onto the electrode surface before imprinting | |
| carbon fibers modified with Fe3O4/MnO2 nanoparticles [32] | Carcinoembryonic Antigen | Self-polymerization | nanosheet | MnO2 nanosheets were incorporated into this design due to their ability to enhance the affinity and biocompatibility of the supports through interactions with proteins | Manageable dimensions and structure, a significant surface area, as well as straightforward surface modification | |
| AuNPs-CNT-MIPda [43] | urea | Electropolymerization | NA | Conductive properties | A covalent bond is established between the numerous AuNPs and the thiol groups present on the aptamer | |
| Gold electrode [42] | Sulfamethoxazole | Electropolymerization | NA | Conductive properties | -- | |
| Carboneous materials | multi-walled carbon nanotubes [44] | sunset yellow | Self-polymerization | The thickness layer of MIP: ~1.8 nm The thickness layer of NIP: ~3.5 nm | Conductive properties | The difference between the thickness layers of MIP and NIP indicates that sunset yellow inhibited the self-polymerization of dopamine, to some extent, on the MWCNT surface |
| Pencil graphite electrode [46] | malathion | Electropolymerization | The roughness of the electrode increased its modification with MIP | Electrode support | In this work, a peptide nanotube (functionalized PDA-based and molecularly imprinted) was employed | |
| Screen printed carbon electrode—Reduced graphene oxide [45] | Epitope of gliadin | Electropolymerization | -- | Reduced graphene oxide enhances the sensitivity of the sensor | Reduced graphene oxide improved the speed of electron transfer kinetics, contributing to a lowered detection limit and an extended linear detection range | |
| Quantum dots | Quantum dots [47] | rabbit IgG | Self-polymerization | QDs-MIPda have spherical shapes as Quantum dots except with a rougher surface | superb optical properties and the ability to multiplex | Dopamine was polymerized in the presence of Caffeic acid to offer carboxyl groups |
| silica NPs | silica NPs [48] | Sunset yellow | Self-polymerization | SiO2 NPs::smooth surface and an average diameter of ~15 nm. The MIPda layer thickness is ~5.5 nm The NIPda layer thickness is ~8.0 nm | supporting matrice to prepare surface imprinted PDA | Dopamine adheres to the surface of SiO2. The difference between the thickness layer of MIPda and NIPda indicates that sunset inhibits the self-polymerization of dopamine, to some extent, on the SiO2 surface |
| Hollow (Silica NPs were removed) [51] | horseradish peroxidase | Self-polymerization | A very thin layer of MIPda | sacrificial matrix | The imprinting factor of the hollow MIP was better than that of the solid MIPs | |
| QCM crystal | QCM crystal [49] | Pepsin, bovine serum albumin, human serum albumin, and lysozyme | Self-polymerization | NA | Support of Quartz crystal microbalance | The protein recognition on MIPda-functionalized QCM crystals depended on both the match between recognition sites and the target protein, as well as non-specific interactions between proteins and the MIPda film |
| QCM crystal [50] | Hepatitis B antigen | Self-polymerization | The bumpy appearance of the MIPda-QCM crystal | Support of Quartz crystal microbalance | -- |

3. Types of Templates
3.1. Ions
3.2. Molecules
3.3. Epitopes
3.4. Proteins
3.5. Viruses and Bacteria
4. Preparation Methods of MIPda
4.1. Self-Polymerization
4.2. Electropolymerization
5. Characteristics of MIPda
5.1. Structure of PDA
5.2. Morphology
5.3. Wettability
6. Applications of MIP-PDA in Sensors
6.1. MIPda-Based Optical Sensors
6.2. MIPda-Based Electrochemical Sensors
6.3. Solid-Phase Extraction Coupled to Sensors
7. Challenges and Limitations
8. Perspectives
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Elfadil, D.; Lamaoui, A.; Della Pelle, F.; Amine, A.; Compagnone, D. Molecularly Imprinted Polymers Combined with Electrochemical Sensors for Food Contaminants Analysis. Molecules 2021, 26, 4607. [Google Scholar] [CrossRef]
- Lamaoui, A.; Cubillana-Aguilera, L.; Gil, M.L.A.; Amine, A.; Palacios-Santander, J.M. Chapter 16:Analytical Applications of Molecularly Imprinted Polymer-Decorated Magnetic Nanoparticles. In Analytical Applications of Functionalized Magnetic Nanoparticles; Royal Society of Chemistry: Cambridge, UK, 2021; pp. 397–428. [Google Scholar]
- BelBruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119. [Google Scholar] [CrossRef]
- Dabrowski, M.; Lach, P.; Cieplak, M.; Kutner, W. Nanostructured Molecularly Imprinted Polymers for Protein Chemosensing. Biosens. Bioelectron. 2018, 102, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Lahcen, A.A.; Surya, S.G.; Beduk, T.; Vijjapu, M.T.; Lamaoui, A.; Durmus, C.; Timur, S.; Shekhah, O.; Mani, V.; Amine, A.; et al. Metal–Organic Frameworks Meet Molecularly Imprinted Polymers: Insights and Prospects for Sensor Applications. ACS Appl. Mater. Interfaces 2022, 14, 49399–49424. [Google Scholar] [CrossRef] [PubMed]
- Lamaoui, A.; Amine, A. Key Advances in MIP-Based Sensors Applied for Cancer and Cardiovas-Cular Biomarkers Detection. Curr. Top. Med. Chem. 2022, 22, 529–548. [Google Scholar] [CrossRef]
- Uzun, L.; Turner, A.P.F. Molecularly-Imprinted Polymer Sensors: Realising Their Potential. Biosens. Bioelectron. 2016, 76, 131–144. [Google Scholar] [CrossRef]
- Beduk, T.; Ait Lahcen, A.; Tashkandi, N.; Salama, K.N. One-Step Electrosynthesized Molecularly Imprinted Polymer on Laser Scribed Graphene Bisphenol a Sensor. Sens. Actuators B Chem. 2020, 314, 128026. [Google Scholar] [CrossRef]
- Lahcen, A.A.; Baleg, A.A.; Baker, P.; Iwuoha, E.; Amine, A. Synthesis and Electrochemical Characterization of Nanostructured Magnetic Molecularly Imprinted Polymers for 17-β-Estradiol Determination. Sens. Actuators B Chem. 2017, 241, 698–705. [Google Scholar] [CrossRef]
- Lamaoui, A.; Karrat, A.; Amine, A. Molecularly Imprinted Polymer Integrated into Paper-Based Analytical Device for Smartphone-Based Detection: Application for Sulfamethoxazole. Sens. Actuators B Chem. 2022, 368, 132122. [Google Scholar] [CrossRef]
- Bossi, A.M. Plastic Antibodies for Cancer Therapy? Nat. Chem. 2020, 12, 111–112. [Google Scholar] [CrossRef]
- Ben Messaoud, N.; Ait Lahcen, A.; Dridi, C.; Amine, A. Ultrasound Assisted Magnetic Imprinted Polymer Combined Sensor Based on Carbon Black and Gold Nanoparticles for Selective and Sensitive Electrochemical Detection of Bisphenol A. Sens. Actuators B Chem. 2018, 276, 304–312. [Google Scholar] [CrossRef]
- Elfadil, D.; Palmieri, S.; Silveri, F.; Della Pelle, F.; Sergi, M.; Del Carlo, M.; Amine, A.; Compagnone, D. Fast Sonochemical Molecularly Imprinted Polymer Synthesis for Selective Electrochemical Determination of Maleic Hydrazide. Microchem. J. 2022, 180, 107634. [Google Scholar] [CrossRef]
- Karim, K.; Lamaoui, A.; Amine, A. Acetazolamide Detection Via Its Competition with Sulfamethoxazole on Molecularly Imprinted Polymer: A Proof-of-Concept. J. Pharm. Biomed. Anal. 2022, 219, 114954. [Google Scholar] [CrossRef]
- Bouvarel, T.; Delaunay, N.; Pichon, V. Selective Extraction of Cocaine from Biological Samples with a Miniaturized Monolithic Molecularly Imprinted Polymer and On-Line Analysis in Nano-Liquid Chromatography. Anal. Chim. Acta 2020, 1096, 89–99. [Google Scholar] [CrossRef]
- Lamaoui, A.; Lahcen, A.A.; García-Guzmán, J.J.; Palacios-Santander, J.M.; Cubillana-Aguilera, L.; Amine, A. Study of Solvent Effect on the Synthesis of Magnetic Molecularly Imprinted Polymers Based on Ultrasound Probe: Application for Sulfonamide Detection. Ultrason. Sonochem. 2019, 58, 104670. [Google Scholar] [CrossRef]
- Lamaoui, A.; Palacios-Santander, J.M.; Amine, A.; Cubillana-Aguilera, L. Fast Microwave-Assisted Synthesis of Magnetic Molecularly Imprinted Polymer for Sulfamethoxazole. Talanta 2021, 232, 122430. [Google Scholar] [CrossRef]
- Bayramoglu, G.; Arica, M.Y. Synthesis of Cr(VI)-Imprinted Poly(4-Vinyl Pyridine-Co-Hydroxyethyl Methacrylate) Particles: Its Adsorption Propensity to Cr(VI). J. Hazard. Mater. 2011, 187, 213–221. [Google Scholar] [CrossRef]
- Li, B.; Li, C.; Jiang, L.; Zeng, Y.; Wang, N. Preparation of Molecularly Imprinted Polymer Based on Calcium Acrylate and Acrylic Acid. Polym. Sci. Ser. B 2022, 64, 176–187. [Google Scholar] [CrossRef]
- Ding, F.; Gao, X.; Huang, X.; Ge, H.; Xie, M.; Qian, J.; Song, J.; Li, Y.; Zhu, X.; Zhang, C. Polydopamine-Coated Nucleic Acid Nanogel for SiRNA-Mediated Low-Temperature Photothermal Therapy. Biomaterials 2020, 245, 119976. [Google Scholar] [CrossRef]
- Liu, C.; Yao, W.; Tian, M.; Wei, J.; Song, Q.; Qiao, W. Mussel-Inspired Degradable Antibacterial Polydopamine/Silica Nanoparticle for Rapid Hemostasis. Biomaterials 2018, 179, 83–95. [Google Scholar] [CrossRef]
- Seddaoui, N.; Amine, A. A Sensitive Colorimetric Immunoassay Based on Poly(Dopamine) Modified Magnetic Nanoparticles for Meat Authentication. LWT 2020, 122, 109045. [Google Scholar] [CrossRef]
- Lee, H.A.; Park, E.; Lee, H. Polydopamine and Its Derivative Surface Chemistry in Material Science: A Focused Review for Studies at KAIST. Adv. Mater. 2020, 32, 1907505. [Google Scholar] [CrossRef]
- Ryu, J.H.; Messersmith, P.B.; Lee, H. Polydopamine Surface Chemistry: A Decade of Discovery. ACS Appl. Mater. Interfaces 2018, 10, 7523–7540. [Google Scholar] [CrossRef]
- Ouyang, R.; Lei, J.; Ju, H.; Xue, Y. A Molecularly Imprinted Copolymer Designed for Enantioselective Recognition of Glutamic Acid. Adv. Funct. Mater. 2007, 17, 3223–3230. [Google Scholar] [CrossRef]
- Ouyang, R.; Lei, J.; Ju, H. Surface Molecularly Imprinted Nanowire for Protein Specific Recognition. Chem. Commun. 2008, 44, 5761–5763. [Google Scholar] [CrossRef] [PubMed]
- Elfadil, D.; Della Pelle, F.; Compagnone, D.; Amine, A. Green Synthesis of Molecularly Imprinted Polymers for Dispersive Magnetic Solid-Phase Extraction of Erythrosine B Associated with Smartphone Detection in Food Samples. Materials 2022, 15, 7653. [Google Scholar] [CrossRef]
- Lamaoui, A.; Palacios-Santander, J.M.; Amine, A.; Cubillana-Aguilera, L. Molecularly Imprinted Polymers Based on Polydopamine: Assessment of Non-Specific Adsorption. Microchem. J. 2021, 164, 106043. [Google Scholar] [CrossRef]
- Yang, Y.-Y.; Li, Y.-T.; Li, X.-J.; Zhang, L.; Kouadio Fodjo, E.; Han, S. Controllable in Situ Fabrication of Portable AuNP/Mussel-Inspired Polydopamine Molecularly Imprinted SERS Substrate for Selective Enrichment and Recognition of Phthalate Plasticizers. Chem. Eng. J. 2020, 402, 125179. [Google Scholar] [CrossRef]
- Yao, G.-H.; Liang, R.-P.; Huang, C.-F.; Wang, Y.; Qiu, J.-D. Surface Plasmon Resonance Sensor Based on Magnetic Molecularly Imprinted Polymers Amplification for Pesticide Recognition. Anal. Chem. 2013, 85, 11944–11951. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Zhang, J.-W.; Wang, C.-Z.; Zhou, L.-D.; Zhang, Q.-H.; Yuan, C.-S. Polydopamine-Coated Magnetic Molecularly Imprinted Polymers with Fragment Template for Identification of Pulsatilla Saponin Metabolites in Rat Feces with UPLC-Q-TOF-MS. J. Agric. Food Chem. 2018, 66, 653–660. [Google Scholar] [CrossRef]
- Lu, H.; Xu, S. Carbon Nanofibers Coated with Fe3O4 Nanoparticles and MnO2 Nanosheets Further Modified with Molecularly Imprinted Polydopamine for Fluorescence Sensing of Carcinoembryonic Antigen. ACS Appl. Nano Mater. 2022, 5, 2532–2540. [Google Scholar] [CrossRef]
- Palladino, P.; Bettazzi, F.; Scarano, S. Polydopamine: Surface Coating, Molecular Imprinting, and Electrochemistry—Successful Applications and Future Perspectives in (Bio)Analysis. Anal. Bioanal. Chem. 2019, 411, 4327–4338. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Miao, L.; Yang, H.; Yu, J.; Xie, Y.; Xu, L.; Song, Y. A Novel Nanoenzyme Based on Fe3O4 Nanoparticles@thionine-Imprinted Polydopamine for Electrochemical Biosensing. Sens. Actuators B Chem. 2017, 253, 108–114. [Google Scholar] [CrossRef]
- Khumsap, T.; Bamrungsap, S.; Thu, V.T.; Nguyen, L.T. Development of Epitope-Imprinted Polydopamine Magnetic Nanoparticles for Selective Recognition of Allergenic Egg Ovalbumin. Chem. Pap. 2022, 76, 6129–6139. [Google Scholar] [CrossRef]
- Chen, J.; Lei, S.; Xie, Y.; Wang, M.; Yang, J.; Ge, X. Fabrication of High-Performance Magnetic Lysozyme-Imprinted Microsphere and Its NIR-Responsive Controlled Release Property. ACS Appl. Mater. Interfaces 2015, 7, 28606–28615. [Google Scholar] [CrossRef]
- Lan, F.; Ma, S.; Yang, Q.; Xie, L.; Wu, Y.; Gu, Z. Polydopamine-Based Superparamagnetic Molecularly Imprinted Polymer Nanospheres for Efficient Protein Recognition. Colloids Surf. B Biointerfaces 2014, 123, 213–218. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Y.; Liu, J.; Xu, Q.; Xiao, H.; Wang, X.; Xu, H.; Zhou, J. Thiol Modified Fe3O4@SiO2 as a Robust, High Effective, and Recycling Magnetic Sorbent for Mercury Removal. Chem. Eng. J. 2013, 226, 30–38. [Google Scholar] [CrossRef]
- Dashtian, K.; Hajati, S.; Ghaedi, M. L-Phenylalanine-Imprinted Polydopamine-Coated CdS/CdSe n-n Type II Heterojunction as an Ultrasensitive Photoelectrochemical Biosensor for the PKU Monitoring. Biosens. Bioelectron. 2020, 165, 112346. [Google Scholar] [CrossRef]
- Sarabaegi, M.; Roushani, M. Rapid and Sensitive Determination of Pseudomonas Aeruginosa by Using a Glassy Carbon Electrode Modified with Gold Nanoparticles and Aptamer-Imprinted Polydopamine. Microchem. J. 2021, 168, 106388. [Google Scholar] [CrossRef]
- Khumsap, T.; Bamrungsap, S.; Thu, V.T.; Nguyen, L.T. Epitope-Imprinted Polydopamine Electrochemical Sensor for Ovalbumin Detection. Bioelectrochemistry 2021, 140, 107805. [Google Scholar] [CrossRef]
- Turco, A.; Corvaglia, S.; Mazzotta, E.; Pompa, P.P.; Malitesta, C. Preparation and Characterization of Molecularly Imprinted Mussel Inspired Film as Antifouling and Selective Layer for Electrochemical Detection of Sulfamethoxazole. Sens. Actuators B Chem. 2018, 255, 3374–3383. [Google Scholar] [CrossRef]
- Yarahmadi, S.; Azadbakht, A.; Derikvand, R.M. Hybrid Synthetic Receptor Composed of Molecularly Imprinted Polydopamine and Aptamers for Impedimetric Biosensing of Urea. Microchim. Acta 2019, 186, 71. [Google Scholar] [CrossRef]
- Yin, Z.-Z.; Cheng, S.-W.; Xu, L.-B.; Liu, H.-Y.; Huang, K.; Li, L.; Zhai, Y.-Y.; Zeng, Y.-B.; Liu, H.-Q.; Shao, Y.; et al. Highly Sensitive and Selective Sensor for Sunset Yellow Based on Molecularly Imprinted Polydopamine-Coated Multi-Walled Carbon Nanotubes. Biosens. Bioelectron. 2018, 100, 565–570. [Google Scholar] [CrossRef]
- Corpuz, A.; Khumsap, T.; Bamrungsap, S.; Thu, V.T.; Nguyen, L.T. Epitope-Imprinted Polydopamine and Reduced Graphene Oxide-Based Sensing Interface for Label-Free Detection of Gliadin. J. Food Compos. Anal. 2023, 117, 105090. [Google Scholar] [CrossRef]
- Yaman, Y.T.; Bolat, G.; Abaci, S.; Saygin, T.B. Peptide Nanotube Functionalized Molecularly Imprinted Polydopamine Based Single-Use Sensor for Impedimetric Detection of Malathion. Anal. Bioanal. Chem. 2022, 414, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, L.; He, Y.; He, Q.; Ma, H. Quantum-Dots-Encoded-Microbeads Based Molecularly Imprinted Polymer. Biosens. Bioelectron. 2016, 77, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Bonyadi, S.; Ghanbari, K. Development of Highly Sensitive and Selective Sensor Based on Molecular Imprinted Polydopamine-Coated Silica Nanoparticles for Electrochemical Determination of Sunset Yellow. Microchem. J. 2021, 167, 106322. [Google Scholar] [CrossRef]
- Lim, H.J.; Saha, T.; Tey, B.T.; Lal, S.K.; Ooi, C.W. Quartz Crystal Microbalance-Based Biosensing of Proteins Using Molecularly Imprinted Polydopamine Sensing Films: Interplay between Protein Characteristics and Molecular Imprinting Effect. Surf. Interfaces 2023, 39, 102904. [Google Scholar] [CrossRef]
- Lim, H.J.; Saha, T.; Tey, B.T.; Tan, W.S.; Hassan, S.S.; Ooi, C.W. Quartz Crystal Microbalance-Based Biosensing of Hepatitis B Antigen Using a Molecularly Imprinted Polydopamine Film. Talanta 2022, 249, 123659. [Google Scholar] [CrossRef]
- Chen, W.; Fu, M.; Zhu, X.; Liu, Q. Protein Recognition by Polydopamine-Based Molecularly Imprinted Hollow Spheres. Biosens. Bioelectron. 2019, 142, 111492. [Google Scholar] [CrossRef]
- Razmi, H.; Dehghanzade, M. Highly Selective and Sensitive Electrochemical Determination of Ni(II) in Real Samples Based on Ion-Imprinted Polymer Technology. Electroanalysis 2020, 32, 198–206. [Google Scholar] [CrossRef]
- Qiao, L.; Gan, N.; Hu, F.; Wang, D.; Lan, H.; Li, T.; Wang, H. Magnetic Nanospheres with a Molecularly Imprinted Shell for the Preconcentration of Diethylstilbestrol. Microchim. Acta 2014, 181, 1341–1351. [Google Scholar] [CrossRef]
- Li, C.; Han, D.; Wu, Z.; Liang, Z.; Han, F.; Chen, K.; Fu, W.; Han, D.; Wang, Y.; Niu, L. Polydopamine-Based Molecularly Imprinted Electrochemical Sensor for the Highly Selective Determination of Ecstasy Components. Analyst 2022, 147, 3291–3297. [Google Scholar] [CrossRef]
- Arabi, M.; Ostovan, A.; Wang, Y.; Mei, R.; Fu, L.; Li, J.; Wang, X.; Chen, L. Chiral Molecular Imprinting-Based SERS Detection Strategy for Absolute Enantiomeric Discrimination. Nat. Commun. 2022, 13, 5757. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Tang, D.; Zhang, Y.; Xu, L.; Liu, K.; Huang, K.; Yin, Z. Specific and Sensitive Detection of Tartrazine on the Electrochemical Interface of a Molecularly Imprinted Polydopamine-Coated PtCo Nanoalloy on Graphene Oxide. Biosensors 2022, 12, 326. [Google Scholar] [CrossRef]
- Wei, J.; Wu, C.; Wu, X.; Wu, L. A Sensitive Electrochemical Bisphenol A Sensor Based on Molecularly Imprinted Polydopamine-Coated Fe3O4 Microspheres. Anal. Sci. 2022, 38, 339–346. [Google Scholar] [CrossRef]
- Miao, J.; Liu, A.; Wu, L.; Yu, M.; Wei, W.; Liu, S. Magnetic Ferroferric Oxide and Polydopamine Molecularly Imprinted Polymer Nanocomposites Based Electrochemical Impedance Sensor for the Selective Separation and Sensitive Determination of Dichlorodiphenyltrichloroethane (DDT). Anal. Chim. Acta 2020, 1095, 82–92. [Google Scholar] [CrossRef]
- Leibl, N.; Duma, L.; Gonzato, C.; Haupt, K. Polydopamine-Based Molecularly Imprinted Thin Films for Electro-Chemical Sensing of Nitro-Explosives in Aqueous Solutions. Bioelectrochemistry 2020, 135, 107541. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Lu, Z.; Liu, T.; Zhao, L.; Ding, F.; Zou, P.; Wang, X.; Zhao, Q.; Rao, H. Molecularly Imprinted Polydopamine Modified with Nickel Nanoparticles Wrapped with Carbon: Fabrication, Characterization and Electrochemical Detection of Uric Acid. Microchim. Acta 2019, 186, 414. [Google Scholar] [CrossRef]
- Yin, Y.; Yan, L.; Zhang, Z.; Wang, J. Magnetic Molecularly Imprinted Polydopamine Nanolayer on Multiwalled Carbon Nanotubes Surface for Protein Capture. Talanta 2015, 144, 671–679. [Google Scholar] [CrossRef]
- Han, W.; Han, X.; Liu, Z.; Zhang, S.; Li, Y.; Lu, J.; Chen, J.; Ou, L.; Fu, G. Facile Modification of Protein-Imprinted Polydopamine Coatings over Nanoparticles with Enhanced Binding Selectivity. Chem. Eng. J. 2020, 385, 123463. [Google Scholar] [CrossRef]
- Lv, P.; Xie, D.; Zhang, Z. Magnetic Carbon Dots Based Molecularly Imprinted Polymers for Fluorescent Detection of Bovine Hemoglobin. Talanta 2018, 188, 145–151. [Google Scholar] [CrossRef]
- Xia, Z.; Lin, Z.; Xiao, Y.; Wang, L.; Zheng, J.; Yang, H.; Chen, G. Facile Synthesis of Polydopamine-Coated Molecularly Imprinted Silica Nanoparticles for Protein Recognition and Separation. Biosens. Bioelectron. 2013, 47, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Lv, S.; Chen, F.; Liu, C.; Cai, C.; Chen, C.; Chen, X. A Resonance Light Scattering Sensor Based on Bioinspired Molecularly Imprinted Polymers for Selective Detection of Papain at Trace Levels. Anal. Chim. Acta 2016, 912, 125–132. [Google Scholar] [CrossRef]
- Yang, B.; Gong, H.; Chen, C.; Chen, X.; Cai, C. A Virus Resonance Light Scattering Sensor Based on Mussel-Inspired Molecularly Imprinted Polymers for High Sensitive and High Selective Detection of Hepatitis A Virus. Biosens. Bioelectron. 2017, 87, 679–685. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, L.; Gong, H.; Chen, C.; Cai, C. A Magnetic Molecularly Imprinted Optical Chemical Sensor for Specific Recognition of Trace Quantities of Virus. RSC Adv. 2018, 8, 32262–32268. [Google Scholar] [CrossRef]
- Zhang, J.; Li, B.; Yue, H.; Wang, J.; Zheng, Y. Highly Selective and Efficient Imprinted Polymers Based on Carboxyl-Functionalized Magnetic Nanoparticles for the Extraction of Gallic Acid from Pomegranate Rind. J. Sep. Sci. 2018, 41, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yan, L.; Zhang, Z.; Wang, J.; Luo, N. Polydopamine-Coated Magnetic Molecularly Imprinted Polymer for the Selective Solid-Phase Extraction of Cinnamic Acid, Ferulic Acid and Caffeic Acid from Radix Scrophulariae Sample: Liquid Chromatography. J. Sep. Sci. 2016, 39, 1480–1488. [Google Scholar] [CrossRef] [PubMed]
- Aktan, M.K.; Coppola, G.A.; Van der Gucht, M.; Yoshioka, T.; Killian, M.S.; Lavigne, R.; Van der Eycken, E.; Steenackers, H.P.; Braem, A. Influence of Polydopamine Functionalization on the Rapid Protein Immobilization by Alternating Current Electrophoretic Deposition. Surf. Interfaces 2022, 34, 102347. [Google Scholar] [CrossRef]
- Iskierko, Z.; Sharma, P.S.; Noworyta, K.R.; Borowicz, P.; Cieplak, M.; Kutner, W.; Bossi, A.M. Selective PQQPFPQQ Gluten Epitope Chemical Sensor with a Molecularly Imprinted Polymer Recognition Unit and an Extended-Gate Field-Effect Transistor Transduction Unit. Anal. Chem. 2019, 91, 4537–4543. [Google Scholar] [CrossRef]
- Du, X.; Li, L.; Li, J.; Yang, C.; Frenkel, N.; Welle, A.; Heissler, S.; Nefedov, A.; Grunze, M.; Levkin, P.A. UV-Triggered Dopamine Polymerization: Control of Polymerization, Surface Coating, and Photopatterning. Adv. Mater. 2014, 26, 8029–8033. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, S.-H.; Oh, I.-K.; Lee, H. Microwave-Accelerated Rapid, Chemical Oxidant-Free, Material-Independent Surface Chemistry of Poly(Dopamine). Small 2017, 13, 1600443. [Google Scholar] [CrossRef] [PubMed]
- Lamaoui, A.; María Palacios-Santander, J.; Amine, A.; Cubillana-Aguilera, L. Computational Approach and Ultrasound Probe–Assisted Synthesis of Magnetic Molecularly Imprinted Polymer for the Electrochemical Detection of Bisphenol A. Mater. Sci. Eng. B Solid-State Mater. Adv. Technol. 2022, 277, 115568. [Google Scholar] [CrossRef]
- Patel, K.; Singh, N.; Yadav, J.; Nayak, J.M.; Sahoo, S.K.; Lata, J.; Chand, D.; Kumar, S.; Kumar, R. Polydopamine Films Change Their Physicochemical and Antimicrobial Properties with a Change in Reaction Conditions. Phys. Chem. Chem. Phys. 2018, 20, 5744–5755. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liang, R.-P.; Wang, X.-N.; Qiu, J.-D. A Norepinephrine Coated Magnetic Molecularly Imprinted Polymer for Simultaneous Multiple Chiral Recognition. J. Chromatogr. A 2015, 1409, 268–276. [Google Scholar] [CrossRef]
- Baldoneschi, V.; Palladino, P.; Banchini, M.; Minunni, M.; Scarano, S. Norepinephrine as New Functional Monomer for Molecular Imprinting: An Applicative Study for the Optical Sensing of Cardiac Biomarkers. Biosens. Bioelectron. 2020, 157, 112161. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.W.; Peterson, M.B.; Ellersdorfer, P.; Granville, A.M. Expanding the Aqueous-Based Redox-Facilitated Self-Polymerization Chemistry of Catecholamines to 5,6-Dihydroxy-1H-Benzimidazole and Its 2-Substituted Derivatives. RSC Adv. 2016, 6, 25203–25214. [Google Scholar] [CrossRef]
- Tretjakov, A.; Syritski, V.; Reut, J.; Boroznjak, R.; Volobujeva, O.; Öpik, A. Surface Molecularly Imprinted Polydopamine Films for Recognition of Immunoglobulin G. Microchim. Acta 2013, 180, 1433–1442. [Google Scholar] [CrossRef]
- Shahdost-fard, F.; Roushani, M. Impedimetric Detection of Trinitrotoluene by Using a Glassy Carbon Electrode Modified with a Gold Nanoparticle@fullerene Composite and an Aptamer-Imprinted Polydopamine. Microchim. Acta 2017, 184, 3997–4006. [Google Scholar] [CrossRef]
- Lian, W.; Liu, S.; Yu, J.; Xing, X.; Li, J.; Cui, M.; Huang, J. Electrochemical Sensor Based on Gold Nanoparticles Fabricated Molecularly Imprinted Polymer Film at Chitosan-Platinum Nanoparticles/Graphene-Gold Nanoparticles Double Nanocomposites Modified Electrode for Detection of Erythromycin. Biosens. Bioelectron. 2012, 38, 163–169. [Google Scholar] [CrossRef]
- Razavipanah, I.; Alipour, E.; Deiminiat, B.; Rounaghi, G.H. A Novel Electrochemical Imprinted Sensor for Ultrasensitive Detection of the New Psychoactive Substance “Mephedrone”. Biosens. Bioelectron. 2018, 119, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Li, H.; Zhao, Y.; Frazer, L.; Qian, B.; Borguet, E.; Ren, F.; Dikin, D.A. Electrical and Mechanical Properties of Poly(Dopamine)-Modified Copper/Reduced Graphene Oxide Composites. J. Mater. Sci. 2017, 52, 11620–11629. [Google Scholar] [CrossRef]
- Dreyer, D.R.; Miller, D.J.; Freeman, B.D.; Paul, D.R.; Bielawski, C.W. Elucidating the Structure of Poly(Dopamine). Langmuir 2012, 28, 6428–6435. [Google Scholar] [CrossRef]
- Shalev, T.; Gopin, A.; Bauer, M.; Stark, R.W.; Rahimipour, S. Non-Leaching Antimicrobial Surfaces through Polydopamine Bio-Inspired Coating of Quaternary Ammonium Salts or an Ultrashort Antimicrobial Lipopeptide. J. Mater. Chem. 2012, 22, 2026–2032. [Google Scholar] [CrossRef]
- Liebscher, J.; Mrówczyński, R.; Scheidt, H.A.; Filip, C.; Hădade, N.D.; Turcu, R.; Bende, A.; Beck, S. Structure of Polydopamine: A Never-Ending Story? Langmuir 2013, 29, 10539–10548. [Google Scholar] [CrossRef]
- Almeida, L.C.; Correia, J.P.; Viana, A.S. Electrochemical and Optical Characterization of Thin Polydopamine Films on Carbon Surfaces for Enzymatic Sensors. Electrochim. Acta 2018, 263, 480–489. [Google Scholar] [CrossRef]
- Liu, X.; Lin, W.; Xiao, P.; Yang, M.; Sun, L.-P.; Zhang, Y.; Xue, W.; Guan, B.-O. Polydopamine-Based Molecular Imprinted Optic Microfiber Sensor Enhanced by Template-Mediated Molecular Rearrangement for Ultra-Sensitive C-Reactive Protein Detection. Chem. Eng. J. 2020, 387, 124074. [Google Scholar] [CrossRef]
- Wang, W.; Wang, R.; Liao, M.; Kidd, M.T.; Li, Y. Rapid Detection of Enrofloxacin Using a Localized Surface Plasmon Resonance Sensor Based on Polydopamine Molecular Imprinted Recognition Polymer. Food Meas. 2021, 15, 3376–3386. [Google Scholar] [CrossRef]
- Roushani, M.; Sarabaegi, M.; Rostamzad, A. Novel Electrochemical Sensor Based on Polydopamine Molecularly Imprinted Polymer for Sensitive and Selective Detection of Acinetobacter Baumannii. J. Iran Chem. Soc. 2020, 17, 2407–2413. [Google Scholar] [CrossRef]
- Tlili, A.; Attia, G.; Khaoulani, S.; Mazouz, Z.; Zerrouki, C.; Yaakoubi, N.; Othmane, A.; Fourati, N. Contribution to the Understanding of the Interaction between a Polydopamine Molecular Imprint and a Protein Model: Ionic Strength and PH Effect Investigation. Sensors 2021, 21, 619. [Google Scholar] [CrossRef]
- Ding, Y.H.; Floren, M.; Tan, W. Mussel-Inspired Polydopamine for Bio-Surface Functionalization. Biosurface Biotribology 2016, 2, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Liu, C.; Chen, Y. Stability of Polydopamine Coatings on Gold Substrates Inspected by Surface Plasmon Resonance Imaging. Langmuir 2018, 34, 3565–3571. [Google Scholar] [CrossRef] [PubMed]




| Template Family | Template | Removal Solution | Analytical Method | Imprinting Factor |
|---|---|---|---|---|
| Ions | Ni2+ [52] | 0.1 M EDTA (pH = 4.5) | Electrochemical method | N/A |
| Molecules | Sulfamethoxazole [42] | 5% of acetic acid | Electrochemical method | 4.84 |
| Diethylstilbestrol [53] | Electrochemical method | |||
| 3,4-Methylenedioxyamphetamine (MDA) and 3,4-methylenedioxymethamphetamine (MDMA) [54] | 0.1 mol L−1 NaOH | Electrochemical method | >2 | |
| Tryptophan [55] | 0.5% (v/v) solution of acetic acid | surface-enhanced Raman scattering | N/A | |
| Tartrazine [56] | Vethanol: Vconcentrated ammonia:Vwater = 7:2:1 | Electrochemical method | No response by NIPda and high signal of MIPda | |
| BPA [57] | 10% acetic acid and 20% acetonitrile | Electrochemical method | 2.35 | |
| BPA [58] | 20% acetonitrile and 3% acetic acid | Electrochemical method | 2.6 | |
| Sunset yellow [48] | Ethanol, concentrated ammonia, and water with volume ratios of 7:2:1 | Electrochemical method | 2.6 | |
| 2,4,6-trinitrotoluene and 1,3,5-trinitroperhydro-1,3,5-triazine [59] | Methanol: aqueous ammonia solution at pH 9 (1:1, v/v) | Electrochemical method | N/A (the NIP-sensor is completely silent and high signal of MIP-sensor) | |
| L-phenylalanine [39] | N/A | Electrochemical method | 17.2 | |
| Uric acid [60] | Methanol/acetic acid (9:1, v/v) | Electrochemical method | N/A | |
| Epitopes | Ovalbumin IgE-binding epitope [41] | 1 M NaOH | Electrochemical method | No response by NIP |
| Gliadin epitope [45] | Acetic acid 3% (v/v) and SDS 1% (w/v) | Electrochemical method | IF of the epitope = 8.68 IF of the target = 3.14 | |
| Proteins | Pepsin [49] | SDS (10%, w/v) and acetic acid (5%, v/v) | Quartz crystal microbalance | 5.78 |
| Carcinoembryonic antigen [32] | 0.1 M HAc containing 5% SDS (w/v) | Fluorescence | 5.5 | |
| Thionine [34] | acetic acid/acetonitrile solution | Electrochemical method | No response by NIP | |
| Rabbit IgG [47] | 0.1 M glycine-HCL buffer (pH 3.5) | Fluorescence | N/A | |
| Human serum albumin [61] | 0.5 M NaCl | N/A | 4.64 | |
| Lysozyme [62] | 1% SDS/3% HAc | N/A | 6.4 | |
| Lysozyme [37] | SDS and acetic acid (0.1%, w/v: 3%, v/v) | N/A | 4.38 | |
| Horseradish peroxidase [51] | 20% acetic acid | N/A | 2.95 (it is not prominent | |
| Bovine hemoglobin [63] | 0.5% triton x-100 | Fluorescence | 4.1 | |
| Bovine hemoglobin [64] | SDS (10%, w/v) and acetic acid (10%, v/v) | N/A | 5.33 | |
| Papain [65] | Acetic acid (5%, v/v) and SDS (10%, w/v) | Resonance light scattering sensor | N/A (MIP exhibited higher light scattering than NIP) | |
| Viruses | Hepatitis B core antigen [50] | Acetic acid (5%, v/v) and SDS (10%, w/v) | Quartz crystal microbalance | N/A (MIP exhibited higher response than NIP) |
| Hepatitis A [66] | Acetic acid (5%, v/v) and SDS (10%, w/v) | Fluorescence | N/A (MIP exhibited higher response than NIP) | |
| Hepatitis A virus [67] | Acetic acid (5%, v/v) and SDS (10%, w/v) | Fluorescence | N/A (MIP exhibited higher response than NIP) |
| Core Materials | Templates | Electrochemical Conditions | Sensors | Comments |
|---|---|---|---|---|
| Au electrode | Immunoglobulin G [79] | −0.45 and +0.55 Vat a scan rate of 50 mV s−1 in PBS buffer Solution | QCM sensor | The igg was immobilized on the electrode surface before the synthesis of MIPda |
| Au electrode | Sulfamethoxazole [42] | Between−0.6 and 0.6 V at a rate of 20 mv s−1 for 60 cycle | Amperometric detection | No significant imprinting and no considerable difference between MIP and NIP |
| GCE modified with aunps@fullerene | An amino-aptamer for 2,4,6-trinitrotoluene [80] | −0.5 to 0.5 V At 20 mV s−1 (13 cycles) | Impedimetric detection | The preparation method is not easy |
| Nickel nanoparticles Wrapped with carbon | Uric acid [60] | −0.6 to 0.6 V for 10 cycles, scan rate 50 mv/s, and the electrolyte was 0.01 M Phosphate buffer solution (pH 7.4) | DPV | The uric acid can be oxidized during electro-polymerization in the potential range of −0.6 to 0.6 V |
| AuNPs/CNTs/GCE | Urea–aptamer complex [43] | −0.5 to 0.5 V vs. Ag/Agcl at a scan rate of 20 mV s−1 (13 cycles) | Impedance Spectroscopy | The urea was immobilized on the surface SH-AuNPs/CNTs/GCE |
| Cds/Zn/Ti substrate | L-phenylalanine [39] | +1.5 V to −1.5 V at 50 mV/s, 20 cycles | Photoelectrochemical | The template can be oxidized during polymerization |
| Au electrode | Carboxylic-acid-based Structural analogs (’dummy’ templates) for nitro-explosives (2,4,6-trinitrotoluene, TNT) And (1,3,5-trinitroperhydro-1,3,5-triazine [59]. | −0.5 V and +0.5 V 0.02 V s−1 for 15 cycles | -- | Dopamine was identified in silico, based on DFT (density functional theory) calculations |
| Aunp-coated screen-printed carbon electrode | Ovalbumin Ige-binding epitope [41] | −0.5 to +1.0 V at a scan rate of 50 mV/s for 10 cycles | DPV | The pH of solution, concentration of template, and number of cycles of electropolymerization were optimized |
| Target Analyte | Detection Method | LOD | Notable Features |
|---|---|---|---|
| C-reactive protein (CRP) [88] | Microfiber interference sensor | 5.8 × 10−10 ng/mL | Low LOD, label-free diagnosis of CRP |
| Carcinoembryonic antigen (CEA) [32] | Fluorescence sensor | 3.5 pg/mL | Sandwich structure, dual signal amplification |
| Enrofloxacin (ENRO) [89] | Localized SPR biosensor | 25–1000 ng/mL | LSPR-based sensor for ENRO residue detection |
| Target Analyte | Electrode Modification Method | Detection Method | LOD | Notable Features |
|---|---|---|---|---|
| MDA and MDMA [54] | Electrochemical polymerization | Differential Pulse Voltammetry | 37 nM for MDA, 54 nM for MDMA | Rapid detection of illicit stimulants |
| Sunset Yellow [48] | Self-polymerized PDA on MWCNTs | Electrochemical response | 1.4 nM | Precise SY detection, MWCNT cavities |
| Nitro-explosives [59] | Electropolymerized MIP films | Cyclic Voltammetry | 0.1 nM–10 nM | Enhanced sensitivity |
| Ovalbumin protein [41] | Imprinted PDA electrode | Differential Pulse Voltammetry | 10.76 nM | Sensitivity for allergenic protein detection |
| Target Analyte | Extraction Method | Detection Method | LOD | Notable Features |
|---|---|---|---|---|
| Cinnamic acid, ferulic acid, caffeic acid [69] | Magnetic imprinted polymers | HPLC analysis | -- | Selective extraction from complex matrix |
| Human serum albumin (HSA) [61] | Magnetic imprinted polymers | HPLC analysis | -- | Protein capture from urine samples |
| Erythrosine B (ERT-B) [27] | Magnetic dispersive SPE | Smartphone-based colorimetric detection | 0.04 mg/L | Selective extraction and colorimetric detection |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamaoui, A.; Lahcen, A.A.; Amine, A. Unlocking the Potential of Molecularly Imprinted Polydopamine in Sensing Applications. Polymers 2023, 15, 3712. https://doi.org/10.3390/polym15183712
Lamaoui A, Lahcen AA, Amine A. Unlocking the Potential of Molecularly Imprinted Polydopamine in Sensing Applications. Polymers. 2023; 15(18):3712. https://doi.org/10.3390/polym15183712
Chicago/Turabian StyleLamaoui, Abderrahman, Abdellatif Ait Lahcen, and Aziz Amine. 2023. "Unlocking the Potential of Molecularly Imprinted Polydopamine in Sensing Applications" Polymers 15, no. 18: 3712. https://doi.org/10.3390/polym15183712
APA StyleLamaoui, A., Lahcen, A. A., & Amine, A. (2023). Unlocking the Potential of Molecularly Imprinted Polydopamine in Sensing Applications. Polymers, 15(18), 3712. https://doi.org/10.3390/polym15183712