Insights into COVID-19 Vaccine Development Based on Immunogenic Structural Proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity
Abstract
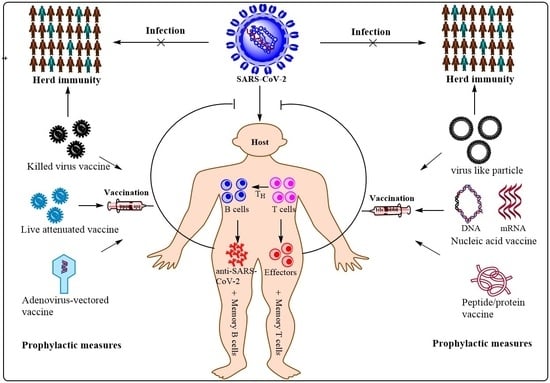
:1. Introduction
2. Nature of Vaccines
2.1. Inactivated or Killed Virus (SARS-CoV-2) Vaccine
2.2. Live-Attenuated Virus (SARS-CoV-2) Vaccine
2.3. Virus-Like Particle (VLP) Vaccine
2.4. Peptide or Protein Subunit Vaccine
2.5. Nucleic Acid (DNA and mRNA) Vaccine
2.6. Virus-Vectored Vaccine
3. Immune Responses to SARS-CoV-2 Structural and Non-Structural Proteins
3.1. Activation of Innate Immune Response against SARS-CoV-2
3.2. Activation of Adaptive (Humoral and Cell-Mediated) Immune Response against SARS-CoV-2
4. Protective Immune Response (Correlates of Protection) against SARS-CoV-2 for Vaccine Development
5. Vaccination, Herd Immunity (Population Immunity) and Herd Immunity Threshold
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- International Committee on Taxonomy of Viruses Executive Committee. The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020, 5, 668–674. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. USA 2020, 117, 26382–26388. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.-S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Eurosurveillance Editorial Team. Note from the editors: World Health Organization declares novel coronavirus (2019-nCoV) sixth public health emergency of international concern. Eurosurveillance 2020, 25, 200131e. [Google Scholar] [CrossRef]
- Arthi, V.; Parman, J. Disease, downturns, and wellbeing: Economic history and the long-run impacts of COVID-19. Explor. Econ. Hist. 2021, 79, 101381. [Google Scholar] [CrossRef]
- To, K.K.-W.; Sridhar, S.; Chiu, K.H.-Y.; Hung, D.L.-L.; Li, X.; Hung, I.F.-N.; Tam, A.R.; Chung, T.W.-H.; Chan, J.F.-W.; Zhang, A.J.-X.; et al. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg. Microbes Infect. 2021, 10, 507–535. [Google Scholar] [CrossRef] [PubMed]
- Kasuga, Y.; Zhu, B.; Jang, K.-J.; Yoo, J.-S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 53, 723–736. [Google Scholar] [CrossRef]
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.; et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021, 184, 2384–2393.e12. [Google Scholar] [CrossRef]
- Gómez, C.; Perdiguero, B.; Esteban, M. Emerging SARS-CoV-2 Variants and Impact in Global Vaccination Programs against SARS-CoV-2/COVID-19. Vaccines 2021, 9, 243. [Google Scholar] [CrossRef]
- Quarleri, J.; Delpino, M.V. Type I and III IFN-mediated antiviral actions counteracted by SARS-CoV-2 proteins and host inherited factors. Cytokine Growth Factor Rev. 2021, 58, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Imran, M.; Dhamija, P.; Chaurasia, D.K.; Handu, S. Virtual screening, ADMET prediction and dynamics simulation of potential compounds targeting the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 39, 1–16. [Google Scholar] [CrossRef]
- Yadav, R.; Hasan, S.; Mahato, S.; Celik, I.; Mary, Y.; Kumar, A.; Dhamija, P.; Sharma, A.; Choudhary, N.; Chaudhary, P.K.; et al. Molecular docking, DFT analysis, and dynamics simulation of natural bioactive compounds targeting ACE2 and TMPRSS2 dual binding sites of spike protein of SARS CoV-2. J. Mol. Liq. 2021, 342, 116942. [Google Scholar] [CrossRef]
- Yadav, R.; Imran, M.; Dhamija, P.; Suchal, K.; Handu, S. Virtual screening and dynamics of potential inhibitors targeting RNA binding domain of nucleocapsid phosphoprotein from SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 39, 4433–4448. [Google Scholar] [CrossRef]
- Yadav, R.; Daman Parihar, R.; Dhiman, U.; Dhamija, P.; Kumar Upadhyay, S.; Imran, M.; Behera, S.K.; Keshava Prasad, T.S. Docking of FDA Approved Drugs Targeting NSP-16, N-Protein and Main Protease of SARS-CoV-2 as Dual Inhibitors. Biointerface Res. Appl. Chem. 2020, 11, 9848–9861. [Google Scholar] [CrossRef]
- Rolta, R.; Yadav, R.; Salaria, D.; Trivedi, S.; Imran, M.; Sourirajan, A.; Baumler, D.J.; Dev, K. In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: An approach to prevent virus assembly. J. Biomol. Struct. Dyn. 2020, 39, 7017–7034. [Google Scholar] [CrossRef]
- Yadav, R.; Chaudhary, J.; Jain, N.; Chaudhary, P.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- Owji, H.; Negahdaripour, M.; Hajighahramani, N. Immunotherapeutic approaches to curtail COVID-19. Int. Immunopharmacol. 2020, 88, 106924. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with COVID-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef]
- Ellah, N.H.A.; Gad, S.F.; Muhammad, K.; Batiha, G.E.; Hetta, H.F. Nanomedicine as a promising approach for diagnosis, treatment and prophylaxis against COVID-19. Nanomedicine 2020, 15, 2085–2102. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, J.K.; Yadav, R.; Chaudhary, P.K.; Maurya, A.; Roshan, R.; Azam, F.; Mehta, J.; Handu, S.; Prasad, R.; Jain, N.; et al. Host Cell and SARS-CoV-2-Associated Molecular Structures and Factors as Potential Therapeutic Targets. Cells 2021, 10, 2427. [Google Scholar] [CrossRef]
- Clemente-Suárez, V.J.; Hormeño-Holgado, A.; Jiménez, M.; Benitez-Agudelo, J.C.; Navarro-Jiménez, E.; Perez-Palencia, N.; Maestre-Serrano, R.; Laborde-Cárdenas, C.C.; Tornero-Aguilera, J.F. Dynamics of Population Immunity Due to the Herd Effect in the COVID-19 Pandemic. Vaccines 2020, 8, 236. [Google Scholar] [CrossRef]
- Plotkin, S.A. Vaccines: The fourth century. Clin. Vaccine Immunol. 2009, 16, 1709–1719. [Google Scholar] [CrossRef] [Green Version]
- Barrett, P.N.; Mundt, W.; Kistner, O.; Howard, M.K. Vero cell platform in vaccine production: Moving towards cell culture-based viral vaccines. Expert Rev. Vaccines 2009, 8, 607–618. [Google Scholar] [CrossRef]
- Shin, M.D.; Shukla, S.; Chung, Y.H.; Beiss, V.; Chan, S.K.; Ortega-Rivera, O.A.; Wirth, D.M.; Chen, A.; Sack, M.; Pokorski, J.K.; et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020, 15, 646–655. [Google Scholar] [CrossRef]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Duan, K.; Zhang, Y.; Zhao, D.; Zhang, H.; Xie, Z.; Li, X.; Peng, C.; Zhang, Y.-B.; Zhang, W.; et al. Effect of an Inactivated Vaccine Against SARS-CoV-2 on Safety and Immunogenicity Outcomes: Interim Analysis of 2 Randomized Clinical Trials. JAMA 2020, 324, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.J. An Inactivated Virus Candidate Vaccine to Prevent COVID-19. JAMA 2020, 324, 943–945. [Google Scholar] [CrossRef]
- Jara, A.; Undurraga, E.A.; González, C.; Paredes, F.; Fontecilla, T.; Jara, G.; Pizarro, A.; Acevedo, J.; Leo, K.; Leon, F.; et al. Effectiveness of an Inactivated SARS-CoV-2 Vaccine in Chile. N. Engl. J. Med. 2021, 385, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Huang, B.; Deng, W.; Quan, Y.; Wang, W.; Xu, W.; Zhao, Y.; Li, N.; Zhang, J.; et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 2020, 182, 713–721.e9. [Google Scholar] [CrossRef] [PubMed]
- Ella, R.; Vadrevu, K.M.; Jogdand, H.; Prasad, S.; Reddy, S.; Sarangi, V.; Ganneru, B.; Sapkal, G.; Yadav, P.; Abraham, P.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: A double-blind, randomised, phase 1 trial. Lancet Infect. Dis. 2021, 21, 637–646. [Google Scholar] [CrossRef]
- Berg, M.K.; Yu, Q.; Salvador, C.E.; Melani, I.; Kitayama, S. Mandated Bacillus Calmette-Guérin (BCG) vaccination predicts flattened curves for the spread of COVID-19. Sci. Adv. 2020, 6, eabc1463. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 nCoV-19 COVID-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681. [Google Scholar] [CrossRef]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatulin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G.; et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- Sadoff, J.; Le Gars, M.; Shukarev, G.; Heerwegh, D.; Truyers, C.; de Groot, A.M.; Stoop, J.; Tete, S.; Van Damme, W.; Leroux-Roels, I.; et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S COVID-19 Vaccine. N. Engl. J. Med. 2021, 384, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Li, Y.-H.; Guan, X.-H.; Hou, L.-H.; Wang, W.-J.; Li, J.-X.; Wu, S.-P.; Wang, B.-S.; Wang, Z.; Wang, L.; et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: A dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.E.; Robert, W.; Frenck, J.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; et al. Safety and Immunogenicity of Two RNA-Based COVID-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Chu, L.; McPhee, R.; Huang, W.; Bennett, H.; Pajon, R.; Nestorova, B.; Leav, B. A preliminary report of a randomized controlled phase 2 trial of the safety and immunogenicity of mRNA-1273 SARS-CoV-2 vaccine. Vaccine 2021, 39, 2791–2799. [Google Scholar] [CrossRef] [PubMed]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2020, 384, 403–416. [Google Scholar] [CrossRef]
- Zhang, N.-N.; Li, X.-F.; Deng, Y.-Q.; Zhao, H.; Huang, Y.-J.; Yang, G.; Huang, W.-J.; Gao, P.; Zhou, C.; Zhang, R.-R.; et al. A Thermostable mRNA Vaccine against COVID-19. Cell 2020, 182, 1271–1283.e16. [Google Scholar] [CrossRef] [PubMed]
- Richmond, P.; Hatchuel, L.; Dong, M.; Ma, B.; Hu, B.; Smolenov, I.; Li, P.; Liang, P.; Han, H.H.; Liang, J.; et al. Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: A phase 1, randomised, double-blind, placebo-controlled trial. Lancet 2021, 397, 682–694. [Google Scholar] [CrossRef]
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef] [PubMed]
- Minor, P.D. Live attenuated vaccines: Historical successes and current challenges. Virology 2015, 479–480, 379–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohn, K.G.-I.; Smith, I.; Sjursen, H.; Cox, R.J. Immune responses after live attenuated influenza vaccination. Hum. Vaccin. Immunother. 2018, 14, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Jang, Y. Cold-Adapted Live Attenuated SARS-Cov-2 Vaccine Completely Protects Human ACE2 Transgenic Mice from SARS-Cov-2 Infection. Vaccines 2020, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Ciapponi, A.; Bardach, A.; Ares, L.R.; Glujovsky, D.; Cafferata, M.L.; Cesaroni, S.; Bhatti, A. Sequential inactivated (IPV) and live oral (OPV) poliovirus vaccines for preventing poliomyelitis. Cochrane Database Syst. Rev. 2019, 12. [Google Scholar] [CrossRef]
- Plotkin, S. History of vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 12283–12287. [Google Scholar] [CrossRef] [Green Version]
- Poland, G.A.; Ovsyannikova, I.G.; Crooke, S.; Kennedy, R.B. SARS-CoV-2 Vaccine Development: Current Status. Mayo Clin. Proc. 2020, 95, 2172–2188. [Google Scholar] [CrossRef]
- SBiering-Sørensen, S.; Aaby, P.; Napirna, B.M.; Roth, A.; Ravn, H.; Rodrigues, A.; Whittle, H.; Benn, C.S. Small randomized trial among low-birth-weight children receiving bacillus Calmette-Guérin vaccination at first health center contact. Pediatr. Infect. Dis. J. 2012, 31, 306–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aaby, P.; Roth, A.; Ravn, H.; Napirna, B.M.; Rodrigues, A.; Lisse, I.M.; Stensballe, L.G.; Diness, B.R.; Lausch, K.R.; Lund, N.; et al. Randomized trial of BCG vaccination at birth to low-birth-weight children: Beneficial nonspecific effects in the neonatal period? J. Infect. Dis. 2011, 204, 245–252. [Google Scholar] [CrossRef]
- Arts, R.J.; Moorlag, S.J.; Novakovic, B.; Li, Y.; Wang, S.-Y.; Oosting, M.; Kumar, V.; Xavier, R.J.; Wijmenga, C.; Joosten, L.A.; et al. BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe 2018, 23, 89–100.e5. [Google Scholar] [CrossRef] [Green Version]
- Blok, B.A.; Arts, R.J.W.; Van Crevel, R.; Benn, C.S.; Netea, M.G. Trained innate immunity as underlying mechanism for the long-term, nonspecific effects of vaccines. J. Leukoc. Biol. 2015, 98, 347–356. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef]
- Kalita, P.; Padhi, A.K.; Zhang, K.Y.J.; Tripathi, T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb. Pathog. 2020, 145, 104236. [Google Scholar] [CrossRef]
- Vogel, F.R.; Sarver, N. Nucleic acid vaccines. Clin. Microbiol. Rev. 1995, 8, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Spier, R. Nucleic acid vaccines. 17-18 May 1994, WHO (OMS), Geneva, Switzerland. Vaccine 1995, 13, 131–132. [Google Scholar] [CrossRef]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; McMahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef]
- Hobernik, D.; Bros, M. DNA Vaccines-How Far from Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef] [Green Version]
- Kutzler, M.A.; Weiner, D.B. DNA vaccines: Ready for prime time? Nat. Rev. Genet. 2008, 9, 776–788. [Google Scholar] [CrossRef]
- Ledwith, B.J.; Manam, S.; Troilo, P.J.; Barnum, A.B.; Pauley, C.J.; Griffiths, T.G.; Harper, L.B.; Beare, C.M.; Bagdon, W.J.; Nichols, W.W. Plasmid DNA vaccines: Investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology 2000, 43, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Mairhofer, J.; Pfaffenzeller, I.; Merz, D.; Grabherr, R. A novel antibiotic free plasmid selection system: Advances in safe and efficient DNA therapy. Biotechnol. J. 2008, 3, 83–89. [Google Scholar] [CrossRef]
- Cranenburgh, R.M.; Hanak, J.A.J.; Williams, S.G.; Sherratt, D.J. Escherichia coli strains that allow antibiotic-free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 2001, 29, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, K.M.; Tzeng, T.-T.; Shen, K.-Y.; Liao, H.-C.; Lin, J.-J.; Chen, M.-Y.; Yu, G.-Y.; Dou, H.-Y.; Liao, C.-L.; Chen, H.-W.; et al. DNA vaccination induced protective immunity against SARS CoV-2 infection in hamsterss. PLoS Negl. Trop. Dis. 2021, 15, e0009374. [Google Scholar] [CrossRef]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New Vaccine Technologies to Combat Outbreak Situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardi, N.; Hogan, M.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Brisse, M.; Vrba, S.M.; Kirk, N.; Liang, Y.; Ly, H. Emerging Concepts and Technologies in Vaccine Development. Front. Immunol. 2020, 11, 2578. [Google Scholar] [CrossRef]
- Sasso, E.; D’Alise, A.M.; Zambrano, N.; Scarselli, E.; Folgori, A.; Nicosia, A. Viruses as vaccine vectors for infectious diseases and cancer. Nat. Rev. Genet. 2009, 8, 62–73. [Google Scholar] [CrossRef]
- Sasso, E.; D’Alise, A.M.; Zambrano, N.; Scarselli, E.; Folgori, A.; Nicosia, A. New viral vectors for infectious diseases and cancer. Semin. Immunol. 2020, 50, 101430. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, J.J.; Sparrer, K.M.J.; Van Gent, M.; Lässig, C.; Huang, T.; Osterrieder, N.; Hopfner, K.-P.; Gack, M.U. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat. Immunol. 2018, 19, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Innate Immunity to Virus Infection. Immunol. Rev. 2009, 227, 75–86. [Google Scholar] [CrossRef]
- Hayn, M.; Hirschenberger, M.; Koepke, L.; Nchioua, R.; Straub, J.H.; Klute, S.; Hunszinger, V.; Zech, F.; Bozzo, C.P.; Aftab, W.; et al. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 2021, 35, 109126. [Google Scholar] [CrossRef]
- Flores-Torres, A.S.; Salinas-Carmona, M.C.; Salinas, E.; Rosas-Taraco, A.G. Eosinophils and Respiratory Viruses. Viral Immunol. 2019, 32, 198–207. [Google Scholar] [CrossRef]
- Li, Y.X.; Wu, W.; Yang, T.; Zhou, W.; Fu, Y.M.; Feng, Q.M.; Ye, J.M. Characteristics of peripheral blood leukocyte differential counts in patients with COVID-19. Zhonghua Nei Ke Za Zhi 2020, 59, 372–374. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Tu, L.; Zhu, P.; Mu, M.; Wang, R.; Yang, P.; Wang, X.; Hu, C.; Ping, R.; Hu, P.; et al. Clinical Features of 85 Fatal Cases of COVID-19 from Wuhan. A Retrospective Observational Study. Am. J. Respir. Crit. Care Med. 2020, 201, 1372–1379. [Google Scholar] [CrossRef] [Green Version]
- Azkur, A.K.; Akdis, M.; Azkur, D.; Sokolowska, M.; Van De Veen, W.; Brüggen, M.-C.; O’Mahony, L.; Gao, Y.; Nadeau, K.; Akdis, C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 2020, 75, 1564–1581. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Coyne, C.B.; Zeh, H.J.; Lotze, M.T. PAMPs and DAMPs: Signal 0s that spur autophagy and immunity. Immunol. Rev. 2012, 249, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L. Complexity of danger: The diverse nature of damage-associated molecular patterns. J. Biol. Chem. 2014, 289, 35237–35245. [Google Scholar] [CrossRef] [Green Version]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370. [Google Scholar] [CrossRef]
- Wang, E.Y.; Team, Y.I.; Mao, T.; Klein, J.; Dai, Y.; Huck, J.D.; Jaycox, J.R.; Liu, F.; Zhou, T.; Israelow, B.; et al. Diverse functional autoantibodies in patients with COVID-19. Nature 2021, 595, 283–288. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nat. Cell Biol. 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; De Jesus, P.D.; et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNab, F.W.; Mayerbarber, K.D.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Rebendenne, A.; Valadão, A.L.C.; Tauziet, M.; Maarifi, G.; Bonaventure, B.; McKellar, J.; Planès, R.; Nisole, S.; Arnaud-Arnould, M.; Moncorgé, O.; et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. 2021, 95, e02415-20. [Google Scholar] [CrossRef]
- Ribero, M.S.; Jouvenet, N.; Dreux, M.; Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020, 16, e1008737. [Google Scholar] [CrossRef]
- Acharya, D.; Liu, G.; Gack, M.U. Dysregulation of type I interferon responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 397–398. [Google Scholar] [CrossRef]
- Schultze, J.L.; Aschenbrenner, A.C. COVID-19 and the human innate immune system. Cell 2021, 184, 1671–1692. [Google Scholar] [CrossRef]
- Gomez, G.N.; Abrar, F.; Dodhia, M.P.; Gonzalez, F.G.; Nag, A. SARS coronavirus protein nsp1 disrupts localization of Nup93 from the nuclear pore complex. Biochem. Cell Biol. 2019, 97, 758–766. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Devaraj, S.G.; Wang, N.; Chen, Z.; Chen, Z.; Tseng, M.; Barretto, N.; Lin, R.; Peters, C.J.; Tseng, C.-T.K.; Baker, S.C.; et al. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007, 282, 32208–32221. [Google Scholar] [CrossRef] [Green Version]
- Frieman, M.; Ratia, K.; Johnston, R.E.; Mesecar, A.; Baric, R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 2009, 83, 6689–6705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagemeijer, M.C.; Monastyrska, I.; Griffith, J.; van der Sluijs, P.; Voortman, J.; Henegouwen, P.M.V.B.E.; Vonk, A.M.; Rottier, P.J.; Reggiori, F.; de Haan, C.A. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology 2014, 458, 125–135. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Wang, D.; Zhou, J.; Pan, T.; Chen, J.; Yang, Y.; Lv, M.; Ye, X.; Peng, G.; Fang, L.; et al. Porcine Deltacoronavirus nsp5 Antagonizes Type I Interferon Signaling by Cleaving STAT2. J. Virol. 2017, 91, e00003-17. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Fang, L.; Wang, D.; Yang, Y.; Chen, J.; Ye, X.; Foda, M.F.F.A.; Xiao, S. Porcine deltacoronavirus nsp5 inhibits interferon-β production through the cleavage of NEMO. Virology 2017, 502, 33–38. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.-C.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Moreira, R.A.; Guzman, H.V.; Boopathi, S.; Baker, J.L.; Poma, A.B. Characterization of Structural and Energetic Differences between Conformations of the SARS-CoV-2 Spike Protein. Materials 2020, 13, 5362. [Google Scholar] [CrossRef]
- Yuan, Y.; Cao, D.; Zhang, Y.; Ma, J.; Qi, J.; Wang, Q.; Lu, G.; Wu, Y.; Yan, J.; Shi, Y.; et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017, 8, 15092. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Padoan, A.; Sciacovelli, L.; Basso, D.; Negrini, D.; Zuin, S.; Cosma, C.; Faggian, D.; Matricardi, P.; Plebani, M. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clin. Chim. Acta 2020, 507, 164–166. [Google Scholar] [CrossRef]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y.; et al. Evolution of antibody immunity to SARS-CoV-2. Nature 2021, 591, 639–644. [Google Scholar] [CrossRef]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Wec, A.Z.; Wrapp, D.; Herbert, A.S.; Maurer, D.; Haslwanter, D.; Sakharkar, M.; Jangra, R.K.; Dieterle, M.E.; Lilov, A.; Huang, D.; et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020, 369, 731–736. [Google Scholar] [CrossRef]
- Barnes, C.O.; West, A.P., Jr.; Huey-Tubman, K.E.; Hoffmann, M.A.G.; Sharaf, N.G.; Hoffman, P.R.; Koranda, N.; Gristick, H.B.; Gaebler, C.; Muecksch, F.; et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. BioRxiv. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Francesco, L. Publisher Correction: COVID-19 antibodies on trial. Nat. Biotechnol. 2021, 39, 246. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Zhou, D.; Ginn, H.M.; Duyvesteyn, H.M.E.; Supasa, P.; Case, J.B.; Zhao, Y.; Walter, T.S.; Mentzer, A.J.; Liu, C.; et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 2021, 184, 2183–2200.e22. [Google Scholar] [CrossRef]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.e16. [Google Scholar] [CrossRef] [PubMed]
- Suryadevara, N.; Shrihari, S.; Gilchuk, P.; VanBlargan, L.A.; Binshtein, E.; Zost, S.J.; Nargi, R.S.; Sutton, R.E.; Winkler, E.S.; Chen, E.C.; et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell 2021, 184, 2316–2331.e15. [Google Scholar] [CrossRef] [PubMed]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371. [Google Scholar] [CrossRef]
- Chen, X.; Pan, Z.; Yue, S.; Yu, F.; Zhang, J.; Yang, Y.; Li, R.; Liu, B.; Yang, X.; Gao, L.; et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct. Target. Ther. 2020, 5, 180. [Google Scholar] [CrossRef] [PubMed]
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef]
- Weiskopf, D.; Schmitz, K.S.; Raadsen, M.P.; Grifoni, A.; Okba, N.M.A.; Endeman, H.; Akker, J.P.C.V.D.; Molenkamp, R.; Koopmans, M.P.G.; van Gorp, E.C.M.; et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020, 5, eabd2071. [Google Scholar] [CrossRef]
- Berger, A. Th1 and Th2 responses: What are they? BMJ 2000, 321, 424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil Melgaço, J.; Brito e Cunha, D.; Azamor, T.; Da Silva, A.M.V.; Tubarão, L.N.; Gonçalves, R.B.; Monteiro, R.Q.; Missailidis, S.; Neves, P.C.D.C.; Bom, A.P.D.A. Cellular and Molecular Immunology Approaches for the Development of Immunotherapies against the New Coronavirus (SARS-CoV-2): Challenges to Near-Future Breakthroughs. J. Immunol. Res. 2020, 2020, 8827670. [Google Scholar] [CrossRef]
- De Paula, C.B.V.; De Azevedo, M.L.V.; Nagashima, S.; Martins, A.P.C.; Malaquias, M.A.S.; Miggiolaro, A.F.R.D.S.; Júnior, J.D.S.M.; Avelino, G.; Carmo, L.A.P.D.; Carstens, L.B.; et al. IL-4/IL-13 remodeling pathway of COVID-19 lung injury. Sci. Rep. 2020, 10, 18689. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fett, C.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014, 88, 11034–11044. [Google Scholar] [CrossRef] [Green Version]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; He, Y.; Jiang, S.; Zheng, B.-J. Development of subunit vaccines against severe acute respiratory syndrome. Drugs Today 2008, 44, 63–73. [Google Scholar] [CrossRef]
- Sun, J.; Zhuang, Z.; Zheng, J.; Li, K.; Wong, R.L.-Y.; Liu, D.; Huang, J.; He, J.; Zhu, A.; Zhao, J.; et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell 2020, 182, 734. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Zhao, M.; Liu, K.; Xu, K.; Wong, G.; Tan, W.; Gao, G.F. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antivir. Res. 2017, 137, 82–92. [Google Scholar] [CrossRef]
- Sariol, A.; Perlman, S. Lessons for COVID-19 Immunity from Other Coronavirus Infections. Immunity 2020, 53, 248–263. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, J.; Mangalam, A.; Channappanavar, R.; Fett, C.; Meyerholz, D.; Agnihothram, S.; Baric, R.S.; David, C.S.; Perlman, S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity 2016, 44, 1379–1391. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Xu, L.; Xie, X.; Yan, H.; Xie, B.; Xu, W.; Liu, X.; Kang, G.; Jiang, W.; Yuan, J. Pulmonary pathology of early-phase COVID-19 pneumonia in a patient with a benign lung lesion. Histopathology 2020, 77, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.A. Correlates of Protection Induced by Vaccination. Clin. Vaccine Immunol. 2010, 17, 1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.-L.; Wang, Z.-Y.; Duan, L.-J.; Meng, Q.-C.; Jiang, M.-D.; Cao, J.; Yao, L.; Zhu, K.-L.; Cao, W.-C.; Ma, M.-J. Susceptibility of Circulating SARS-CoV-2 Variants to Neutralization. N. Engl. J. Med. 2021, 384, 2354–2356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, G.; Pan, H.; Li, C.; Hu, Y.; Chu, K.; Han, W.; Chen, Z.; Tang, R.; Yin, W.; et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021, 21, 181–192. [Google Scholar] [CrossRef]
- Emary, K.R.W.; Golubchik, T.; Aley, P.K.; Ariani, C.V.; Angus, B.; Bibi, S.; Blane, B.; Bonsall, D.; Cicconi, P.; Charlton, S.; et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): An exploratory analysis of a randomised controlled trial. Lancet 2021, 397, 1351–1362. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Lambe, T.; Spencer, A.; Belij-Rammerstorfer, S.; Purushotham, J.N.; Port, J.R.; Avanzato, V.A.; Bushmaker, T.; Flaxman, A.; Ulaszewska, M.; et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020, 586, 578–582. [Google Scholar] [CrossRef]
- Ramasamy, M.N.; Minassian, A.M.; Ewer, K.J.; Flaxman, A.L.; Folegatti, P.M.; Owens, D.R.; Voysey, M.; Aley, P.K.; Angus, B.; Babbage, G.; et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet 2020, 396, 1979–1993. [Google Scholar] [CrossRef]
- Gushchin, V.; Dolzhikova, I.; Shchetinin, A.; Odintsova, A.; Siniavin, A.; Nikiforova, M.; Pochtovyi, A.; Shidlovskaya, E.; Kuznetsova, N.; Burgasova, O.; et al. Neutralizing Activity of Sera from Sputnik V-Vaccinated People against Variants of Concern (VOC: B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.617.3) and Moscow Endemic SARS-CoV-2 Variants. Vaccines 2021, 9, 779. [Google Scholar] [CrossRef]
- Mercado, N.B.; Zahn, R.; Wegmann, F.; Loos, C.; Chandrashekar, A.; Yu, J.; Liu, J.; Peter, L.; McMahan, K.; Tostanoski, L.H.; et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 2020, 586, 583–588. [Google Scholar] [CrossRef]
- Tostanoski, L.H.; Wegmann, F.; Martinot, A.J.; Loos, C.; McMahan, K.; Mercado, N.B.; Yu, J.; Chan, C.N.; Bondoc, S.; Starke, C.E.; et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 2020, 26, 1694–1700. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.-C.; Guan, X.-H.; Li, Y.-H.; Huang, J.-Y.; Jiang, T.; Hou, L.-H.; Li, J.-X.; Yang, B.-F.; Wang, L.; Wang, W.-J.; et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Xia, H.; Zhang, X.; Zou, J.; Fontes-Garfias, C.R.; Weaver, S.C.; Swanson, K.A.; Cai, H.; Sarkar, R.; et al. BNT162b2-Elicited Neutralization against New SARS-CoV-2 Spike Variants. N. Engl. J. Med. 2021, 385, 472–474. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Anderson, R.M.; May, R.M. Vaccination and herd immunity to infectious diseases. Nature 1985, 318, 323–329. [Google Scholar] [CrossRef]
- Randolph, H.E.; Barreiro, L.B. Herd Immunity: Understanding COVID-19. Immunity 2020, 52, 737. [Google Scholar] [CrossRef] [PubMed]
- Plans-Rubió, P. Are the Objectives Proposed by the WHO for Routine Measles Vaccination Coverage and Population Measles Immunity Sufficient to Achieve Measles Elimination from Europe? Vaccines 2020, 8, 184–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallinga, J.; Lipsitch, M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc. R. Soc. B Biol. Sci. 2007, 274, 599–604. [Google Scholar] [CrossRef] [Green Version]
- Nishiura, H.; Linton, N.M.; Akhmetzhanov, A.R. Serial interval of novel coronavirus (COVID-19) infections. Int. J. Infect. Dis. 2020, 93, 284–286. [Google Scholar] [CrossRef] [PubMed]
- Kwok, K.O.; Lai, F.; Wei, W.I.; Wong, S.Y.S.; Tang, J.W.T. Herd immunity–estimating the level required to halt the COVID-19 epidemics in affected countries. J. Infect. 2020, 80, e32–e33. [Google Scholar] [CrossRef]
- Bernal, J.L.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef] [PubMed]




| Brand Name | Current Dose/Gap and Route of Administration | Primary Developer(s) | Country/NRA | Clinical Trial Phase and Identifier | Approved/ Under Development | Reported Efficacy | Ref. |
|---|---|---|---|---|---|---|---|
| A. Inactivated or killed virus (SARS-CoV-2) vaccine (produced in Vero cells) | |||||||
| CoronaVac (formerly PiCoVacc) | Two doses, between 14 and 18 days apart, intramuscular | Sinovac Biotech | China/NMPA | 1 (NCT04352608) 2 (NCT04383574) 3 (NCT04456595) | Approved | Phase 3; 65.9% | [32] |
| BBIBP-CorV | Two doses, intramuscular injection | Sinopharm, Beijing Institute of Biological Products Co. Ltd. | China/NMPA | 1 Not found 2 (ChiCTR2000032459) 3 (ChiCTR2000034780) | Approved | Phase 3; 86% | [33] |
| WIBP-CorV | Two doses, intramuscular injection | Wuhan Institute of Biological Products; China National Pharmaceutical Group (Sinopharm) | China/NMPA | 1/2 (ChiCTR2000031809) Phase 3 trial is awaited | Approved | Phase 1/2; 72.5% | [30] |
| Covaxin (BBV152) | Two doses, 14 days apart, intramuscular | Bharat Biotech, Indian Council of Medical Research (ICMR), National Institute of Virology (NIV) | India/DCGI | 1/2 (NCT04471519) Phase 3 trial is underway | Approved | Interim phase 3; 78% | [34] |
| CoviVac | Not specified | Chumakov Federal Scientific Center for the Research and Development of Immune and Biological Products of the Russian Academy of Sciences | Russia/Russian NRA | Phase 1/2 trial is underway | Approved | Not yet reported | Not yet |
| QazVac (QazCovid-in) | Two doses, 21 days apart, intramuscular | Research Institute for Biological Safety Problems | Kazakhstan | 1/2 (NCT04530357) Phase 3 trial is underway | Approved | 96% | Not yet |
| B. Live-attenuated vaccine against SARS-CoV-2 | |||||||
| Bacillus Calmette-Guerin (BCG) vaccine | Single dose, intradermally | University of Melbourne and Murdoch Children’s Research Institute; Radboud University Medical Center; Faustman Lab at Massachusetts General Hospital | Multinational | 1 (NCT04328441) 2/3 (NCT04327206) | Not yet approved; under development | Not yet known | [35] |
| C. Adenovirus vector-based recombinant vaccine * (Recombinant ChAdOx1 adenoviral vector encoding the SARS-CoV-2 spike protein antigen) # (Human recombinant Adenovirus Vector (rAd5-S or rAd26-S) encoding the SARS-CoV-2 spike protein antigen) $ (Recombinant, replication-incompetent adenovirus type 26 (rAd26) vectored vaccine encoding the SARS-CoV-2 spike protein antigen) @ (Human recombinant Adenovirus Vector (rAd5-S) encoding the SARS-CoV-2 spike protein antigen) | |||||||
| * COVID-19 Vaccine AstraZeneca/ (AZD1222) Vaxzevria/ Covishield | Two doses, between 4 and 12 weeks apart, intramuscular injection | AstraZeneca, University of Oxford, Serum Institute of India | United Kingdom (UK)/ EMA | 1/2 (NCT04324606) 2/3 (NCT04400838) 3 NCT04516746 | Approved | 79% efficacy in phase 3 clinical trial (NCT04516746); 100% efficacy in severe disease and hospitalization patients | [36,37] |
| # Sputnik V (formerly Gam-COVID-Vac Lyo) (rAd5-S or rAd26-S) | Two doses, 21 days apart, intramuscular injection | Gamaleya Research Institute, Acellena Contract Drug Research and Development | Russia/Russian NRA | 1/2 (NCT04436471) and (NCT04436471) 3 (NCT04530396) | Approved | 91.6% efficacy in phase 3 clinical trial | [38,39] |
| # Sputnik light vaccine (rAd26-S) | No. of doses and gap are not yet finalized, intramuscular injection | Gamaleya Research Institute, Acellena Contract Drug Research and Development | Russia/Russian NRA | 1/2 (NCT04713488) 3 (NCT04741061) | Approved | 79.4% efficacy in phase 3 clinical trial | Not yet |
| $ COVID-19 Vaccine Janssen (JNJ-78436735; Ad26.COV2.S) | Single dose vaccine, intramuscular injection | Janssen vaccines (Johnsons & Johnsons) | The Netherlands, US/EMA | 1/2 (NCT04436276) 3 (NCT04505722) | Approved | 85% efficacy in phase 3 ENSEMBLE trial | [40,41] |
| @ Convidicea (Ad5-nCoV) | Single dose vaccine, but also evaluated in trial with 2 doses, intramuscular | CanSino Biologics | China/EMPA | 1 (NCT04313127) 2 (NCT04341389) 3 (NCT04526990) | Approved | 65.7% efficiency in interim phase 3 clinical trial | [42] |
| D. mRNA vaccine (BNT162b2 is a lipid nanoparticle–formulated, nucleoside-modified mRNA vaccine encodes prefusion spike protein) (mRNA-1273 encodes the prefusion-stabilized S protein of SARS-CoV-2) (ARCoV: lipid nanoparticle-encapsulated mRNA (mRNA-LNP) encodes the receptor-binding domain (RBD) of SARS-CoV-2) | |||||||
| Comirnaty (formerly BNT162b2) | Two doses, 21 days apart, intramuscular injection | Pfizer, BioNTech; Fosun Pharma | Multinational/EMA | 1/2 (NCT04380701) 2 (NCT04649021) 2/3 (NCT04368728) | Approved | ~90% efficacy in phase 3 clinical trail | [43,44] |
| Moderna COVID-19 Vaccine (mRNA-1273) | Two doses, 28 days apart, intramuscular injection | Moderna, BARDA, NIAID | The USA/EMA | 1 (NCT04283461) 2 (NCT04405076) 3 (NCT04470427) | Approved | ~94.1% efficacy in phase 3 clinical trial | [45,46] |
| ARCoV | Intramuscular injection | Academy of Military Medical Sciences, Walvax Biotechnology, Suzhou Abogen Biosciences | China/NMPA | ChiCTR2000034112 | Under development | Not yet reported | [47] |
| E. peptide/subunit Vaccine | |||||||
| EpiVacCorona | Two doses, 21–28 days apart, intramuscular injection | Federal Budgetary Research Institution State Research Center of Virology and Biotechnology | Russia/Russian NRA | 1/2 (NCT04527575) 3 (NCT04780035) | Approved | Not yet reported | Not yet |
| SCB-2019(stabilized trimeric form of the spike (S)-protein (S-Trimer) | Two doses, 21 days apart, intramuscular | Glaxo SmithKline, Sanofi, Clover Biopharmaceuticals, Dynavax and Xiamen Innovax | Australia | 1 (NCT04405908) 2/3 is underway | Under development | Not yet reported | [48] |
| F. DNA Vaccine (Plasmid DNA expressing S protein) | |||||||
| INO-4800 | Two doses, intradermal injection | INOVIO Pharmaceuticals, International Vaccine Institute | USA | 1 (NCT04336410) 2/3 (NCT04642638) | Under development | Not yet specified | [49] |
| AG0301-COVID-19 | Two doses, 14 days apart, intramuscular injection | AnGes, Inc. | Japan | 1/2 (NCT04463472) | Under development | Not yet specified | Not yet |
| GX-19N | Two doses, 29 days apart, intramuscular injection | Genexine | South Korea | 1/2a (NCT04715997) | Under development | Not yet specified | Not yet |
| CORVax12 | Two doses, 28 days apart, DNA electroporation | OncoSec; Providence Cancer Institute | The USA | 1 (NCT04627675) | Under development | Not yet specified | Not yet |
| G. Virus-like particle (VLP) or nanoparticle vaccine | |||||||
| ABNCoV2 | Two doses, 28 days apart, intramuscular injection | ExpreS2ion Biotech; Bavarian Nordic A/S | Netherlands | 1 (NCT04839146) | Under development | Not yet specified | Not yet |
| SpFN (spike ferritin nanoparticle vaccine) | Doses and gap are unspecified, intramuscular injection | US Army Medical Research and Development Command | The USA | 1 (NCT04784767) | Under development | Not yet specified | Not yet |
| Name of Structural Proteins | Length (Amino Acids) | Predicted Average AntiGenic Propensity Score | NCBI Ref. Sequence |
|---|---|---|---|
| Spike (S) glycoprotein | 1273 | 1.0146 | YP_009724390.1 |
| Membrane (M) glycoprotein | 222 | 1.0532 | YP_009724393.1 |
| Envelope (E) protein | 75 | 1.1202 | YP_009724392.1 |
| Nucleocapsid (N) phosphoprotein | 419 | 0.9871 | YP_009724397.2 |
| Vaccine | Humoral Response (IgG) (Wild-type SARS-CoV-2) | Cellular Response (Wild type-SARS-CoV-2) | Reported Effectiveness against SARS-CoV-2 Variants of Concern (VOC) | Ref. |
|---|---|---|---|---|
| CoronaVac (formerly PiCoVacc) | Induction of specific IgG against S and N proteins, RBD in mice, rats, and non-human primates (pre-clinical); induction of anti-RBD IgG and nAbs in humans (Clinical) | No detectable induction of T cell response (TH1 or TH2) cell responses in NHPs as well as human | Effective against D614G, and B.1.1.7 Less effective against B.1.351 | [29,143,144] |
| BBIBP-CorV | Induction of nAbs in mice, rats, rabbits, guinea pigs, NHPs (Macaca fascicularis and Rhesus macaques), and humans | No induction of either TH1 or TH2 cell responses in NHPs | Effective against B.1.1.7 Less effective against B.1.351 | [33,34] |
| WIBP-CorV | Formation of virus-specific IgG and nAbs in humans | No report of specific induction of either TH1 or TH2 cell responses in NHPs | Not yet known/reported | [30] |
| Covaxin (BBV152) | Neutralizing antibody (nAbs) response in humans | T cell responses, with biasness towards TH1 cells | Effective against B.1.1.7; effective against B.1.617 | [34] |
| COVID-19 Vaccine AstraZeneca/ (AZD1222) Vaxzevria/ Covishield | Induction of anti-S antibody and nAbs in mice, NHPs, as well as humans, with nAb titres similar to convalescent plasma | Induction of high TH1 cell, but low TH2 cell responses in mice | Reduced neutralisation activity against the B.1.1.7 variant in vitro; however, effective against B.1.1.7 in vivo | [145,146,147] |
| Sputnik V (formerly Gam-COVID-Vac Lyo) (rAd5-S or rAd26-S) | Induction of both RBD-specific antibody and nAbs in humans | Induction of TH and Tc cell responses | Significant neutralizing activity against B.1.1.7, B.1.351, P.1, B.1.617.2 and B.1.617.3 | [39,148] |
| COVID-19 Vaccine Janssen (JNJ-78436735; Ad26.COV2.S) | Generation of both RBD-specific and neutralizing antibodies in hamsters and NHPs | Induction of high TH1, but low TH2 cell responses in NHPs | Effective against B.1.617.2 | [40,41,149,150] |
| Convidicea (Ad5-nCoV) | Generation of RBD-specific and neutralizing antibodies in humans | Generation of TH1 cell response | Not yet known/reported | [42,151] |
| Comirnaty (formerly BNT162b2) | Generation of RBD-specific and neutralizing antibodies (nAbs) in humans | Not yet known | Effective against B.1.526, B.1.429 and B.1.1.7 variants | [43,152] |
| Moderna COVID-19 Vaccine (mRNA-1273) | Generation of S-specific and nAbs in mice, NHPs, and humans | Induction of high TH1, but low TH2 cell responses in mice, NHPs and human | Effective against B.1.351 and P.1 variants; this vaccine also neutralizes the B.1.617.1 variant, albeit 6.8-fold less effectively | [45,153,154] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaudhary, J.K.; Yadav, R.; Chaudhary, P.K.; Maurya, A.; Kant, N.; Rugaie, O.A.; Haokip, H.R.; Yadav, D.; Roshan, R.; Prasad, R.; et al. Insights into COVID-19 Vaccine Development Based on Immunogenic Structural Proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity. Cells 2021, 10, 2949. https://doi.org/10.3390/cells10112949
Chaudhary JK, Yadav R, Chaudhary PK, Maurya A, Kant N, Rugaie OA, Haokip HR, Yadav D, Roshan R, Prasad R, et al. Insights into COVID-19 Vaccine Development Based on Immunogenic Structural Proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity. Cells. 2021; 10(11):2949. https://doi.org/10.3390/cells10112949
Chicago/Turabian StyleChaudhary, Jitendra Kumar, Rohitash Yadav, Pankaj Kumar Chaudhary, Anurag Maurya, Nimita Kant, Osamah Al Rugaie, Hoineiting Rebecca Haokip, Deepika Yadav, Rakesh Roshan, Ramasare Prasad, and et al. 2021. "Insights into COVID-19 Vaccine Development Based on Immunogenic Structural Proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity" Cells 10, no. 11: 2949. https://doi.org/10.3390/cells10112949
APA StyleChaudhary, J. K., Yadav, R., Chaudhary, P. K., Maurya, A., Kant, N., Rugaie, O. A., Haokip, H. R., Yadav, D., Roshan, R., Prasad, R., Chatrath, A., Singh, D., Jain, N., & Dhamija, P. (2021). Insights into COVID-19 Vaccine Development Based on Immunogenic Structural Proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity. Cells, 10(11), 2949. https://doi.org/10.3390/cells10112949