Evolution of a Multiple Sex-Chromosome System by Three-Sequential Translocations among Potential Sex-Chromosomes in the Taiwanese Frog Odorrana swinhoana
Abstract
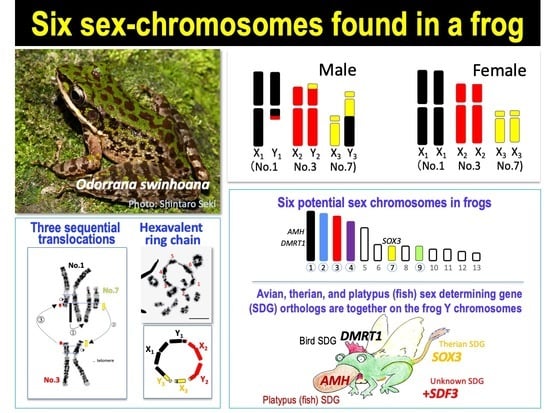
:1. Introduction
2. Materials and Methods
2.1. Frogs
2.2. Chromosome Preparation and Banding Techniques
2.3. Microdissection and Chromosome Painting Probes Preparation
2.4. Telomere Mapping
2.5. Comparative Genomic Hybridization (CGH)
3. Results
3.1. Three Heteromorphic Sex Chromosomes in Males
3.2. A Hexavalent Ring at Male Meiosis
3.3. Direct Proof of the Chromosome Members Involved in Three Translocations
3.4. No Male DNA Specialization on the Three Y Chromosomes
3.5. Population Variation in the Sex-Chromosome System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Graves, J.A.M. The origin and function of the mammalian Y chromosome and Y-borne genes—An evolving understanding. BioEssays 1995, 17, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Graves, J. Review: Sex chromosome evolution and the expression of sex-specific genes in the Placenta. Placenta 2010, 31, S27–S32. [Google Scholar] [CrossRef]
- Kitano, J.; Ross, J.A.; Mori, S.; Kume, M.; Jones, F.C.; Chan, Y.F.; Absher, D.M.; Grimwood, J.; Schmutz, J.; Myers, R.M.; et al. A role for a neo-sex chromosome in stickleback speciation. Nat. Cell Biol. 2009, 461, 1079–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.A.S.; Gordon, I.J.; Traut, W.; Herren, J.; Collins, S.; Martins, D.J.; Saitoti, K.; Ireri, P.; Ffrench-Constant, R. A neo-W chromosome in a tropical butterfly links colour pattern, male-killing, and speciation. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, T.; Kitano, J. The Intricate relationship between sexually antagonistic selection and the evolution of sex chro-mosome fusions. J. Theor. Biol. 2016, 404, 97–108. [Google Scholar] [CrossRef]
- Graves, J.A.M. Sex chromosome specialization and degeneration in mammals. Cell 2006, 124, 901–914. [Google Scholar] [CrossRef] [Green Version]
- Murata, C.; Yamada, F.; Kawauchi, N.; Matsuda, Y.; Kuroiwa, A. The Y chromosome of the Okinawa spiny rat, Tokudaia muenninki, was rescued through fusion with an autosome. Chromosom. Res. 2011, 20, 111–125. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S. The Story of Ancestors—Unfinished: Genes and the Mysteries of Human Origin; Yodosha, Co, Ltd.: Tokyo, Japan, 2000; pp. 1–190. [Google Scholar]
- Morescalchi, M. Amphibia. In Cytotaxanomy and Vertebrate Evolution; Chiarelli, A.B., Capanna, E., Eds.; Academic Press: London, UK; New York, NY, USA, 1973; pp. 233–348. [Google Scholar]
- Miura, I. The late replication banding patterns of chromosomes are highly conserved in the genera Rana, Hyla, and Bufo (Amphibia: Anura). Chromosoma 1995, 103, 567–574. [Google Scholar] [CrossRef]
- Miura, I.; Nishioka, M.; Borkin, L.J.; Wu, Z. The origin of the brown frogs with 2n = 24 chromosomes. Cell. Mol. Life Sci. 1995, 51, 179–188. [Google Scholar] [CrossRef]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.-L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Sex determination: Why so many ways of doing It? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [Green Version]
- Kuramoto, M. Karyotypes of several frogs from Korea, Taiwan and the Philippines. Cell. Mol. Life Sci. 1980, 36, 826–828. [Google Scholar] [CrossRef]
- Kuramoto, M. Karyotypes of the Rana narina complex (Anura: Ranidae) from Japan and Taiwan; Unviersity Bulletin: Fukuoka, Japan, 1983; Volume 45, pp. 27–35. [Google Scholar]
- Schmid, M.; Steinlein, C.; Feichtinger, W. Chromosome banding in amphibia XVII. First demonstration of multiple sex chro-mosomes in amphibians: Eleutherodactylus maussi (Anura, Leptodactylidae). Chromosoma 1992, 101, 284–292. [Google Scholar] [CrossRef]
- Schmid, M.; Feichtinger, W.; Steinlein, C.; García, R.V.; Badillo, A.F. Chromosome banding in Amphibia. XXVIII. Homomorphic XY sex chromosomes and a derived Y-autosome translocation in Eleutherodactylus riveroi (Anura, Leptodactylidae). Cytogenet. Genome Res. 2003, 101, 62–73. [Google Scholar] [CrossRef]
- Schmid, M.; Steinlein, C.; Bogart, J.; Feichtinger, W.; León, P.; La Marca, E.; Diaz, L.; Sanz, A.; Chen, S.-H.-; Hedges, S. The chromosomes of Terraranan frogs. Insights into vertebrate cytogenetics. Cytogenet. Genome Res. 2010, 130–131, 1–14. [Google Scholar] [CrossRef]
- Schmid, M.; Steinlein, C. Chromosome banding in Amphibia. XXXVII. Y-autosome translocations in Anura. Cytogenet. Genome Res. 2018, 154, 153–180. [Google Scholar] [CrossRef] [PubMed]
- Gazoni, T.; Haddad, C.F.B.; Narimatsu, H.; Cabral-De-Mello, D.C.; Lyra, M.L.; Parise-Maltempi, P.P. More sex chromosomes than autosomes in the Amazonian frog Leptodactylus pentadactylus. Chromosom 2018, 127, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, X.; Xia, Y.; Zeng, X. Suppressed recombination of sex chromosomes is not caused by chromosomal reciprocal transloca-tion in spiny frog (Quasipaa boulengeri). Front. Genet. 2018, 9, 288. [Google Scholar] [CrossRef]
- Miura, I. Sex determination and sex chromosomes in Amphibia. Sex. Dev. 2017, 11, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, D.L.; Lavanchy, G.; Sermier, R.; Sredl, M.J.; Miura, I.; Borzée, A.; Barrow, L.N.; Canestrelli, D.; Crochet, P.-A.; Dufresnes, C.; et al. A rapid rate of sex-chromosome turnover and non-random transitions in true frogs. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Miura, I. Sex chromosome differentiation in the Japanese brown frog, Rana japonica I. Sex-related heteromorphism of the dis-tribution pattern of constitutive heterochromatin in chromosome No.4 of the Wakuya population. Zool. Sci. 1994, 11, 797–806. [Google Scholar]
- Sumner, A. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Takayama, S.; Taniguchi, T.; Iwashita, Y. Application of the 4Na-EDTA Giemsa staining method for analysis of DNA repli-cation Chromosome Information Service. CIS 1981, 31, 36–38. [Google Scholar]
- Ezaz, T.; Quinn, A.E.; Miura, I.; Sarre, S.D.; Georges, A.; Graves, J.A.M. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosom. Res. 2005, 13, 763–776. [Google Scholar] [CrossRef]
- Matsubara, K.; Sarre, S.D.; Georges, A.; Matsuda, Y.; Graves, J.A.M.; Ezaz, T. Highly differentiated ZW sex microchromosomes in the Australian Varanus species evolved through rapid amplification of repetitive sequences. PLoS ONE 2014, 9, e95226. [Google Scholar] [CrossRef]
- Yang, F.; Trifonov, V.; Ng, B.L.; Kosyakova, N.; Carter, N.P. Generation of paint probes by flow-sorted and microdissected chromosomes. In Fluorescence In Situ Hybridization (FISH)—Application Guide; Springer: Berlin, Germania, 2009; Volume 1, pp. 35–52. [Google Scholar]
- Zwick, M.S.; Hanson, R.E.; Islam-Faridi, M.N.; Stelly, D.M.; Wing, R.A.; Price, H.J.; McKnight, T.D. A rapid procedure for the isolation of Cot-1 DNA from plants. Genome 1997, 40, 138–142. [Google Scholar] [CrossRef] [Green Version]
- Yano, C.F.; Bertollo, L.A.C.; Cioffi, M.D.B. Fish-FISH: Molecular cytogenetics in fish species. In Immunity in Insects; Springer: Berlin, Germania, 2016; pp. 429–443. [Google Scholar]
- Matsuda, Y.; Chapman, V.M. Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 1995, 16, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Srikulnath, K.; Matsubara, K.; Uno, Y.; Thongpan, A.; Suputtitada, S.; Apisitwanich, S.; Matsuda, Y.; Nishida, C. Karyological characterization of the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Squamata) by molecular cytogenetic approach. Cytogenet. Genome Res. 2009, 125, 213–223. [Google Scholar] [CrossRef]
- Pennell, M.W.; Kirkpatrick, M.; Otto, S.P.; Vamosi, J.C.; Peichel, C.L.; Valenzuela, N.; Kitano, J. Y Fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 2015, 11, e1005237. [Google Scholar] [CrossRef] [Green Version]
- Miura, I. An Evolutionary witness: The frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex. Dev. 2007, 1, 323–331. [Google Scholar] [CrossRef]
- Smith, C.A.; Roeszler, K.N.; Ohnesorg, T.; Cummins, D.M.; Farlie, P.G.; Doran, T.J.; Sinclair, A.H. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nat. Cell Biol. 2009, 461, 267–271. [Google Scholar] [CrossRef]
- Cortez, D.; Marin, R.; Toledo-Flores, D.; Froidevaux, L.; Liechti, A.; Waters, P.D.; Grützner, F.; Kaessmann, H. Origins and functional evolution of Y chromosomes across mammals. Nat. Cell Biol. 2014, 508, 488–493. [Google Scholar] [CrossRef]
- Hattori, R.S.; Murai, Y.; Oura, M.; Masuda, S.; Majhi, S.K.; Sakamoto, T.; Fernandino, J.I.; Somoza, G.M.; Yokota, M.; Strüssmann, C.A. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 2012, 109, 2955–2959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uno, Y.; Nishida, C.; Oshima, Y.; Yokoyama, S.; Miura, I.; Matsuda, Y.; Nakamura, M. Comparative chromosome mapping of sex-linked genes and identification of sex chromosomal rearrangements in the Japanese wrinkled frog (Rana rugosa, Ranidae) with ZW and XY sex chromosome systems. Chromosom. Res. 2008, 16, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Kodama, M.; Suda, M.; Sakamoto, D.; Iwasaki, T.; Matsuo, Y.; Uno, Y.; Matsuda, Y.; Nakamura, Y.; Maekawa, S.; Katsu, Y.; et al. Molecular cloning and Characterization of anti-müllerian hormone (AMH) from the Japanese wrinkled Frog, Rana rugosa. Endocrinology 2015, 156, 1914–1923. [Google Scholar] [CrossRef] [Green Version]
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.-M.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nat. Cell Biol. 1990, 346, 240–244. [Google Scholar] [CrossRef] [Green Version]
- Berta, P.; Hawkins, J.B.; Sinclair, A.H.; Taylor, A.; Griffiths, B.L.; Goodfellow, P.N.; Fellous, M. Genetic evidence equating SRY and the testis-determining factor. Nat. Cell Biol. 1990, 348, 448–450. [Google Scholar] [CrossRef]
- Takehana, Y.; Matsuda, M.; Myosho, T.; Suster, M.L.; Kawakami, K.; Shin-I, T.; Kohara, Y.; Kuroki, Y.; Toyoda, A.; Fujiyama, A.; et al. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 2014, 5, 4157. [Google Scholar] [CrossRef] [Green Version]
- Toups, M.A.; Rodrigues, N.; Perrin, N.; Kirkpatrick, M. A reciprocal translocation radically reshapes sex-linked inheritance in the common frog. Mol. Ecol. 2019, 28, 1877–1889. [Google Scholar] [CrossRef] [PubMed]
- Sigeman, H.; Ponnikas, S.; Chauhan, P.; Dierickx, E.; Brooke, M.D.L.; Hansson, B. Repeated sex chromosome evolution in vertebrates supported by expanded avian sex chromosomes. Proc. R. Soc. B Biol. Sci. 2019, 286, 20192051. [Google Scholar] [CrossRef]
- Singh, L.; Phillips, C.; Jones, K.W. The conserved nucleotide sequences of Bkm, which define Sxr in the mouse, are transcribed. Cell 1984, 36, 111–120. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Augstenová, B.; Clemente, L.; Auer, M.; Frit, U.; Praschag, P.; Protiva, T.; Velenský, P.; Kratochvíl, L.; Rovatsos, M. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae). Sci. Rep. 2020, 10, 4276. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miura, I.; Shams, F.; Lin, S.-M.; de Bello Cioffi, M.; Liehr, T.; Al-Rikabi, A.; Kuwana, C.; Srikulnath, K.; Higaki, Y.; Ezaz, T. Evolution of a Multiple Sex-Chromosome System by Three-Sequential Translocations among Potential Sex-Chromosomes in the Taiwanese Frog Odorrana swinhoana. Cells 2021, 10, 661. https://doi.org/10.3390/cells10030661
Miura I, Shams F, Lin S-M, de Bello Cioffi M, Liehr T, Al-Rikabi A, Kuwana C, Srikulnath K, Higaki Y, Ezaz T. Evolution of a Multiple Sex-Chromosome System by Three-Sequential Translocations among Potential Sex-Chromosomes in the Taiwanese Frog Odorrana swinhoana. Cells. 2021; 10(3):661. https://doi.org/10.3390/cells10030661
Chicago/Turabian StyleMiura, Ikuo, Foyez Shams, Si-Min Lin, Marcelo de Bello Cioffi, Thomas Liehr, Ahmed Al-Rikabi, Chiao Kuwana, Kornsorn Srikulnath, Yuya Higaki, and Tariq Ezaz. 2021. "Evolution of a Multiple Sex-Chromosome System by Three-Sequential Translocations among Potential Sex-Chromosomes in the Taiwanese Frog Odorrana swinhoana" Cells 10, no. 3: 661. https://doi.org/10.3390/cells10030661
APA StyleMiura, I., Shams, F., Lin, S. -M., de Bello Cioffi, M., Liehr, T., Al-Rikabi, A., Kuwana, C., Srikulnath, K., Higaki, Y., & Ezaz, T. (2021). Evolution of a Multiple Sex-Chromosome System by Three-Sequential Translocations among Potential Sex-Chromosomes in the Taiwanese Frog Odorrana swinhoana. Cells, 10(3), 661. https://doi.org/10.3390/cells10030661