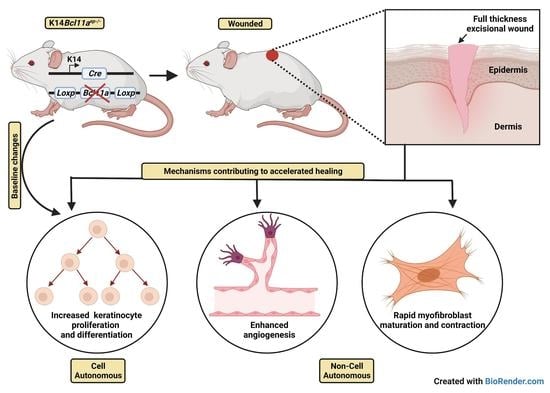
Selective Ablation of BCL11A in Epidermal Keratinocytes Alters Skin Homeostasis and Accelerates Excisional Wound Healing In Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Tail DNA Isolation and Polymerase Chain Reaction (PCR) Genotyping
2.3. Murine Tail Skin Epidermis-Dermis Separation
2.4. Wound Healing Assay
2.5. Immunoblotting
2.6. Histology
2.7. Immunohistochemistry (IHC)
2.8. Imaging
2.9. IHC Analysis and Quantification
2.10. Immunoblot Analysis and Quantification
2.11. Statistical Analysis
3. Results
3.1. Confirmation of Epidermis Specific Bcl11a Excision in Bcl11aep−/− Mice
3.2. Altered Epidermal Homeostasis in Bcl11aep−/− Adult Skin
3.3. Accelerated Cutaneous Wound Healing in Bcl11aep−/− Mice
3.4. Bcl11a in a Cell Autonomous Manner Promotes Accelerated Healing via Early Keratinocyte Activation, Rapid Re-Epithelialization and Onset of Differentiation
3.5. Epidermal Bcl11a Deletion Promotes Advanced Angiogenesis and Myofibroblast Mediated Wound Contraction in a Non-Cell Autonomous Manner
3.6. Rapid Re-Establishment of Epidermal Homeostasis in the Later Stages of Healing in Bcl11aep−/− Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biggs, L.C.; Kim, C.S.; Miroshnikova, Y.A.; Wickstrom, S.A. Mechanical forces in the skin: Roles in tissue architecture, stability, and function. J. Investig. Dermatol. 2020, 140, 284–290. [Google Scholar] [CrossRef]
- Nejati, R.; Kovacic, D.; Slominski, A. Neuro-immune-endocrine functions of the skin: An overview. Expert Rev. Dermatol. 2013, 8, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Berumen Sánchez, G.; Bunn, K.E.; Pua, H.H.; Rafat, M. Extracellular vesicles: Mediators of intercellular communication in tissue injury and disease. Cell Commun. Signal. 2021, 19, 104. [Google Scholar] [CrossRef]
- Alonso, L.; Fuchs, E. Stem cells of the skin epithelium. Proc. Natl. Acad. Sci. USA 2003, 100 (Suppl. S1), 11830–11835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rognoni, E.; Watt, F.M. Skin cell heterogeneity in development, wound healing, and cancer. Trends Cell Biol. 2018, 28, 709–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaels, B. Skin Sampling Techniques: Handbook of Topical Antimicrobials and Their Applications; Paulson, D., Ed.; Marcel Dekker Inc.: New York, NY, USA, 2002; Chapter 26, pp. 395–410. [Google Scholar]
- Gonzales, K.A.U.; Fuchs, E. Skin and its regenerative powers: An alliance between stem cells and their niche. Dev. Cell 2017, 43, 387–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watt, F.M. Mammalian skin cell biology: At the interface between laboratory and clinic. Science 2014, 346, 937–940. [Google Scholar] [CrossRef] [PubMed]
- Joost, S.; Zeisel, A.; Jacob, T.; Sun, X.; La Manno, G.; Lonnerberg, P.; Linnarsson, S.; Kasper, M. Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 2016, 3, 221–237.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, H.N.; Hardman, M.J. Wound healing: Cellular mechanisms and pathological outcomes. Open Biol. 2020, 10, 200223. [Google Scholar] [CrossRef] [PubMed]
- Bielefeld, K.A.; Amini-Nik, S.; Alman, B.A. Cutaneous wound healing: Recruiting developmental pathways for regeneration. Cell Mol. Life Sci. 2013, 70, 2059–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haensel, D.; Jin, S.; Sun, P.; Cinco, R.; Dragan, M.; Nguyen, Q.; Cang, Z.; Gong, Y.; Vu, R.; MacLean, A.L.; et al. Defining Epidermal Basal Cell States during Skin Homeostasis and Wound Healing Using Single-Cell Transcriptomics. Cell Rep. 2020, 30, 3932–3947.e6. [Google Scholar] [CrossRef]
- Guo, S.; Dipietro, L.A. Factors affecting wound healing. J. Dent. Res. 2010, 89, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Martin, P. Wound healing—Aiming for perfect skin regeneration. Science 1997, 276, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Kosaric, N.; Bonham, C.A.; Gurtner, G.C. Wound healing: A cellular perspective. Physiol. Rev. 2019, 99, 665–706. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.J.; Clark, R.A. Cutaneous wound healing. N. Engl. J. Med. 1999, 341, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Braiman-Wiksman, L.; Solomonik, I.; Spira, R.; Tennenbaum, T. Novel insights into wound healing sequence of events. Toxicol. Pathol. 2007, 35, 767–779. [Google Scholar] [CrossRef]
- Guerrero-Juarez, C.F.; Dedhia, P.H.; Jin, S.; Ruiz-Vega, R.; Ma, D.; Liu, Y.; Yamaga, K.; Shestova, O.; Gay, D.L.; Yang, Z.; et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 2019, 10, 650. [Google Scholar] [CrossRef]
- Bellavia, G.; Fasanaro, P.; Melchionna, R.; Capogrossi, M.C.; Napolitano, M. Transcriptional control of skin reepithelialization. J. Dermatol. Sci. 2014, 73, 3–9. [Google Scholar] [CrossRef]
- Boudra, R.; Ramsey, M.R. Understanding transcriptional networks regulating initiation of cutaneous wound healing. Yale J. Biol. Med. 2020, 93, 161–173. [Google Scholar]
- Liang, X.; Bhattacharya, S.; Bajaj, G.; Guha, G.; Wang, Z.; Jang, H.S.; Leid, M.; Indra, A.K. and Ganguli-Indra, G. Delayed cutaneous wound healing and aberrant expression of hair follicle stem cell markers in mice selectively lacking Ctip2 in epidermis. PLoS ONE 2012, 7, e29999. [Google Scholar] [CrossRef] [Green Version]
- Schafer, M.; Werner, S. Transcriptional control of wound repair. Annu. Rev. Cell Dev. Biol. 2007, 23, 69–92. [Google Scholar] [CrossRef]
- Avram, D.; Fields, A.; Pretty On Top, K.; Nevrivy, D.J.; Ishmael, J.E.; Leid, M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 2000, 275, 10315–10322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leid, M.; Ishmael, J.; Avram, D.; Shepherd, D.; Fraulob, V.; Dollé, P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr. Patterns 2004, 4, 733–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, J.; Xie, X.; Ye, Y.; Wang, L.; Che, F. BCL11A: A potential diagnostic biomarker and therapeutic target in human diseases. Biosci. Rep. 2019, 39, BSR20190604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, R.; Wiegreffe, C.; Britsch, S. Bcl11 Transcription factors regulate cortical development and function. Front. Mol. Neurosci. 2020, 13, 51. [Google Scholar] [CrossRef] [Green Version]
- Satterwhite, E.; Sonoki, T.; Willis, T.G.; Harder, L.; Nowak, R.; Arriola, E.L.; Liu, H.; Price, H.P.; Gesk, S.; Steinemann, D.; et al. The BCL11 gene family: Involvement of BCL11A in lymphoid malignancies. Blood 2001, 98, 3413–3420. [Google Scholar] [CrossRef] [Green Version]
- Kliewer, S.A.; Umesono, K.; Heyman, R.A.; Mangelsdorf, D.J.; Dyck, J.A.; Evans, R.M. Retinoid X receptor-COUP-TF interactions modulate retinoic acid signaling. Proc. Natl. Acad. Sci. USA 1992, 89, 1448–1452. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Teegarden, A.; Bauer, E.M.; Choi, J.; Messaddeq, N.; Hendrix, D.A.; Ganguli-Indra, G.; Leid, M.; Indra, A.K. Transcription Factor CTIP1/ BCL11A regulates epidermal differentiation and lipid metabolism during skin development. Sci. Rep. 2017, 7, 13427. [Google Scholar] [CrossRef] [Green Version]
- Dassule, H.R.; Lewis, P.; Bei, M.; Maas, R.; McMahon, A.P. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 2000, 127, 4775–4785. [Google Scholar] [CrossRef]
- DeLisser, H.M.; Newman, P.J.; Albelda, S.M. Platelet endothelial cell adhesion molecule (CD31). Curr. Top. Microbiol. Immunol. 1993, 184, 37–45. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, L.J.; Guha, G.; Li, S.; Kyrylkova, K.; Kioussi, C.; Leid, M.; Ganguli-Indra, G.; Indra, A.K. Selective ablation of Ctip2/Bcl11b in epidermal keratinocytes triggers atopic dermatitis-like skin inflammatory responses in adult mice. PLoS ONE 2012, 7, e51262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Zieman, A.; Coulombe, P.A. Skin Keratins. Methods Enzymol. 2016, 568, 303–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousselle, P.; Gentilhomme, E.; Neveux, Y. Agache's Measuring the Skin: Non-Invasive Investigations, Physiology, Normal Constants; Humbert, P., Fanian, F., Maibach, H.I., Agache, P., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 407–415. [Google Scholar]
- Pastar, I.; Stojadinovic, O.; Yin, N.C.; Ramirez, H.; Nusbaum, A.G.; Sawaya, A.; Patel, S.B.; Khalid, L.; Isseroff, R.R.; Tomic-Canic, M. Epithelialization in wound healing: A comprehensive review. Adv. Wound Care 2014, 3, 445–464. [Google Scholar] [CrossRef] [Green Version]
- Florin, L.; Knebel, J.; Zigrino, P.; Vonderstrass, B.; Mauch, C.; Schorpp-Kistner, M.; Szabowski, A.; Angel, P. Delayed wound healing and epidermal hyperproliferation in mice lacking JunB in the skin. J. Investig. Dermatol. 2006, 126, 902–911. [Google Scholar] [CrossRef] [Green Version]
- Patel, G.K.; Wilson, C.H.; Harding, K.G.; Finlay, A.Y.; Bowden, P.E. Numerous keratinocyte subtypes involved in wound re-epithelialization. J. Investig. Dermatol. 2006, 126, 497–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leigh, I.M.; Navsaria, H.; Purkis, P.E.; McKay, I.A.; Bowden, P.E.; Riddle, P.N. Keratins (K16 and K17) as markers of keratinocyte hyperproliferation in psoriasis in vivo and in vitro. Br. J. Dermatol. 1995, 133, 501–511. [Google Scholar] [CrossRef]
- Bigliardi-Qi, M.; Gaveriaux-Ruff, C.; Zhou, H.; Hell, C.; Bady, P.; Rufli, T.; Kieffer, B.; Bigliardi, P.L. Deletion of δ-opioid receptor in mice alters skin differentiation and delays wound healing. Differentiation 2006, 74, 174–185. [Google Scholar] [CrossRef]
- Gerritsen, M.J.; Elbers, M.E.; de Jong, E.M.; van de Kerkhof, P.C. Recruitment of cycling epidermal cells and expression of filaggrin, involucrin and tenascin in the margin of the active psoriatic plaque, in the uninvolved skin of psoriatic patients and in the normal healthy skin. J. Dermatol. Sci. 1997, 14, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Paladini, R.D.; Takahashi, K.; Bravo, N.S.; Coulombe, P.A. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: Defining a potential role for keratin 16. J. Cell Biol. 1996, 132, 381–397. [Google Scholar] [CrossRef] [Green Version]
- Koyama, S.; Purk, A.; Kaur, M.; Soini, H.; Novotny, M.; Davis, K.; Kao, C.; Matsunami, H.; Mescher, A. Beta-caryophyllene enhances wound healing through multiple routes. PLoS ONE 2019, 14, e0216104. [Google Scholar] [CrossRef] [Green Version]
- Rakita, A.; Nikolić, N.; Mildner, M.; Matiasek, J.; Elbe-Bürger, A. Re-epithelialization and immune cell behaviour in an ex vivo human skin model. Sci. Rep. 2020, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Rhim, J.; Jang, I.-S.; Kwon, S.-T.; Song, K.; Yeo, E.-J.; Park, S. Activation of Wound Healing in Aged Rats by Altering the Cellular Mitogenic Potential. J. Gerontology. Ser. A Biol. Sci. Med. Sci. 2010, 65, 704–711. [Google Scholar] [CrossRef] [PubMed]
- Usui, M.L.; Underwood, R.A.; Mansbridge, J.N.; Muffley, L.A.; Carter, W.G.; Olerud, J.E. Morphological evidence for the role of suprabasal keratinocytes in wound reepithelialization. Wound Repair Regen. 2005, 13, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Yufit, T.; Carson, P.; Fiore, D.; Falanga, J.; Lin, X.; Mamakos, L.; Falanga, V. Differential keratin expression during epiboly in a wound model of bioengineered skin and in human chronic wounds. Int. J. Low. Extrem. Wounds 2011, 10, 122–129. [Google Scholar] [CrossRef] [Green Version]
- Stojadinovic, O.; Brem, H.; Vouthounis, C.; Lee, B.; Fallon, J.; Stallcup, M.; Merchant, A.; Galiano, R.D.; Tomic-Canic, M. Molecular pathogenesis of chronic wounds: The role of beta-catenin and c-myc in the inhibition of epithelialization and wound healing. Am. J. Pathol. 2005, 167, 59–69. [Google Scholar] [CrossRef]
- Darby, I.A.; Laverdet, B.; Bonte, F.; Desmouliere, A. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 2014, 7, 301–311. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Wang, J.H. Fibroblasts and myofibroblasts in wound healing: Force generation and measurement. J. Tissue Viability 2011, 20, 108–120. [Google Scholar] [CrossRef] [Green Version]
- Volksdorf, T.; Heilmann, J.; Eming, S.A.; Schawjinski, K.; Zorn-Kruppa, M.; Ueck, C.; Vidal, Y.S.S.; Windhorst, S.; Jucker, M.; Moll, I.; et al. Tight junction proteins claudin-1 and occludin are important for cutaneous wound healing. Am. J. Pathol. 2017, 187, 1301–1312. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhou, L.; Zhang, D.; Tang, W.J.; Tang, D.; Shi, X.L.; Yang, Y.; Zhou, L.; Liu, F.; Yu, Y.; et al. BCL11A promotes the progression of laryngeal squamous cell carcinoma. Front. Oncol. 2020, 10, 375. [Google Scholar] [CrossRef]
- Sunami, Y.; Yokoyama, T.; Yoshino, S.; Takahara, T.; Yamazaki, Y.; Harada, H.; Nakamura, T. BCL11A promotes myeloid leukemogenesis by repressing PU.1 target genes. Blood Adv. 2022, 6, 1827–1843. [Google Scholar] [CrossRef]
- Seachrist, D.D.; Hannigan, M.M.; Ingles, N.N.; Webb, B.M.; Weber-Bonk, K.L.; Yu, P.; Bebek, G.; Singh, S.; Sizemore, S.T.; Varadan, V.; et al. The transcriptional repressor BCL11A promotes breast cancer metastasis. J. Biol. Chem. 2020, 295, 11707–11719. [Google Scholar] [CrossRef] [PubMed]
- Raja Sivamani, K.; Garcia, M.S.; Isseroff, R.R. Wound re-epithelialization: Modulating keratinocyte migration in wound healing. Front. Biosci. 2007, 12, 2849–2868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vagnozzi, A.N.; Reiter, J.F.; Wong, S.Y. Hair follicle and interfollicular epidermal stem cells make varying contributions to wound regeneration. Cell Cycle 2015, 14, 3408–3417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojcik, S.M.; Bundman, D.S.; Roop, D.R. Delayed wound healing in keratin 6a knockout mice. Mol. Cell. Biol. 2020, 20, 5248–5255. [Google Scholar] [CrossRef] [Green Version]
- Donati, G.; Rognoni, E.; Hiratsuka, T.; Liakath-Ali, K.; Hoste, E.; Kar, G.; Kayikci, M.; Russell, R.; Kretzschmar, K.; Mulder, K.W.; et al. Wounding induces dedifferentiation of epidermal Gata6(+) cells and acquisition of stem cell properties. Nat. Cell Biol. 2017, 19, 603–613. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.E.; Wilgus, T.A. Vascular endothelial growth factor and angiogenesis in the regulation of cutaneous wound repair. Adv. Wound Care 2014, 3, 647–661. [Google Scholar] [CrossRef] [Green Version]
- Mills, S.J.; Cowin, A.J.; Kaur, P. Pericytes, mesenchymal stem cells and the wound healing process. Cells 2013, 2, 621–634. [Google Scholar] [CrossRef] [Green Version]
- Tai, Y.; Woods, E.L.; Dally, J.; Kong, D.; Steadman, R.; Moseley, R.; Midgley, A.C. Myofibroblasts: Function, formation, and scope of molecular therapies for skin fibrosis. Biomolecules 2021, 11, 1095. [Google Scholar] [CrossRef]
- Gustems, M.; Woellmer, A.; Rothbauer, U.; Eck, S.H.; Wieland, T.; Lutter, D.; Hammerschmidt, W. c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res. 2014, 42, 3059–3072. [Google Scholar] [CrossRef]
- Luo, G.; Jing, X.; Yang, S.; Peng, D.; Dong, J.; Li, L.; Reinach, P.S.; Yan, D. DNA Methylation Regulates Corneal Epithelial Wound Healing by Targeting miR-200a and CDKN2B. Investig. Ophthalmol. Vis. Sci. 2019, 60, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Morgunova, E.; Jolma, A.; Kaasinen, E.; Sahu, B.; Khund-Sayeed, S.; Das, P.K.; Kivioja, T.; Dave, K.; Zhong, F.; et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 2017, 356, eaaj2239. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Ziff, E.B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science 1991, 251, 186–189. [Google Scholar] [CrossRef]
- Gawronska-Kozak, B.; Grabowska, A.; Kur-Piotrowska, A.; Kopcewicz, M. Foxn1 transcription factor regulates wound healing of skin through promoting epithelial-mesenchymal transition. PLoS ONE 2016, 11, e0150635. [Google Scholar] [CrossRef] [Green Version]
- Segre, J.A.; Bauer, C.; Fuchs, E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999, 22, 356–360. [Google Scholar] [CrossRef] [PubMed]
- De Guzman Strong, C.; Wertz, P.W.; Wang, C.; Yang, F.; Meltzer, P.S.; Andl, T.; Millar, S.E.; Ho, I.C.; Pai, S.Y.; Segre, J.A. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J. Cell Biol. 2006, 175, 661–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koster, M.I.; Dai, D.; Marinari, B.; Sano, Y.; Costanzo, A.; Karin, M.; Roop, D.R. p63 induces key target genes required for epidermal morphogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 3255–3260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wurm, S.; Zhang, J.; Guinea-Viniegra, J.; Garcia, F.; Munoz, J.; Bakiri, L.; Ezhkova, E.; Wagner, E.F. Terminal epidermal differentiation is regulated by the interaction of Fra-2/AP-1 with Ezh2 and ERK1/2. Genes Dev. 2015, 29, 144–156. [Google Scholar] [CrossRef] [Green Version]
- Lewis, C.J.; Mardaryev, A.N.; Sharov, A.A.; Fessing, M.Y.; Botchkarev, V.A. The epigenetic regulation of wound healing. Adv. Wound Care 2014, 3, 468–475. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, N.; Ganguli-Indra, G.; Indra, A.K. CTIP2 and lipid metabolism: Regulation in skin development and associated diseases. Expert Rev. Proteom. 2021, 18, 1009–1017. [Google Scholar] [CrossRef]
- Zhang, L.J.; Bhattacharya, S.; Leid, M.; Ganguli-Indra, G.; Indra, A.K. Ctip2 is a dynamic regulator of epidermal proliferation and differentiation by integrating EGFR and Notch signaling. J. Cell Sci. 2012, 125, 5733–5744. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Kirkwood, J.S.; Taylor, A.W.; Stevens, J.F.; Leid, M.; Ganguli-Indra, G.; Indra, A.K. Transcription factor Ctip2 controls epidermal lipid metabolism and regulates expression of genes involved in sphingolipid biosynthesis during skin development. J. Investig. Dermatol. 2013, 133, 668–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, S.; Wheeler, H.; Leid, M.; Ganguli-Indra, G.; Indra, A.K. Transcription factor CTIP2 maintains hair follicle stem cell pool and contributes to altered expression of LHX2 and NFATC1. J. Investig. Dermatol. 2015, 135, 2593–2602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Kular, L.; Vij, M.; Herter, E.K.; Li, X.; Wang, A.; Chu, T.; Toma, M.A.; Zhang, L.; Liapi, E.; et al. Human skin long noncoding RNA WAKMAR1 regulates wound healing by enhancing keratinocyte migration. Proc. Natl. Acad. Sci. USA 2019, 116, 9443–9452. [Google Scholar] [CrossRef] [Green Version]
- Kuai, L.; Jiang, J.S.; Li, W.; Li, B.; Yin, S.Y. Long non-coding RNAs in diabetic wound healing: Current research and clinical relevance. Int. Wound J. 2022, 19, 583–600. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, W.; Lu, S.; Ma, Z. Modulation of the wound healing through noncoding RNA interplay and GSK-3beta/NF-kappaB signaling interaction. Int. J. Genomics 2021, 2021, 9709290. [Google Scholar] [CrossRef] [PubMed]
- Luan, A.; Hu, M.S.; Leavitt, T.; Brett, E.A.; Wang, K.C.; Longaker, M.T.; Wan, D.C. Noncoding RNAs in wound healing: A new and vast frontier. Adv. Wound Care 2018, 7, 19–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Wu, W.; Zheng, L.; Lin, X.; Tai, Y.; Wang, Y.; Wang, L. Roles of MicroRNA-21 in Skin Wound Healing: A Comprehensive Review. Front. Pharmacol. 2022, 13, 828627. [Google Scholar] [CrossRef]
- Cameron, D.J.; Tong, Z.; Yang, Z.; Kaminoh, J.; Kamiyah, S.; Chen, H.; Zeng, J.; Chen, Y.; Luo, L.; Zhang, K. Essential role of Elovl4 in very long chain fatty acid synthesis, skin permeability barrier function, and neonatal survival. Int. J. Biol. Sci. 2007, 3, 111–119. [Google Scholar] [CrossRef]
- Vasireddy, V.; Uchida, Y.; Salem, N., Jr.; Kim, S.Y.; Mandal, M.N.; Reddy, G.B.; Bodepudi, R.; Alderson, N.L.; Brown, J.C.; Hama, H.; et al. Loss of functional ELOVL4 depletes very long-chain fatty acids (> or =C28) and the unique omega-O-acylceramides in skin leading to neonatal death. Hum. Mol. Genet. 2007, 16, 471–482. [Google Scholar] [CrossRef]
- Li, S.; Fang, X.D.; Wang, X.Y.; Fei, B.Y. Fos-like antigen 2 (FOSL2) promotes metastasis in colon cancer. Exp. Cell Res. 2018, 373, 57–61. [Google Scholar] [CrossRef]
- Belguise, K.; Kersual, N.; Galtier, F.; Chalbos, D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 2005, 24, 1434–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milde-Langosch, K.; Janke, S.; Wagner, I.; Schroder, C.; Streichert, T.; Bamberger, A.M.; Janicke, F.; Loning, T. Role of Fra-2 in breast cancer: Influence on tumor cell invasion and motility. Breast Cancer Res. Treat. 2008, 107, 337–347. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Mai, J.; Li, Y.; Chen, L.; Xu, H.; Zhu, X.; Pan, Q. miR-597 inhibits breast cancer cell proliferation, migration and invasion through FOSL2. Oncol. Rep. 2017, 37, 2672–2678. [Google Scholar] [CrossRef] [Green Version]
- Sarode, P.; Zheng, X.; Giotopoulou, G.A.; Weigert, A.; Kuenne, C.; Gunther, S.; Friedrich, A.; Gattenlohner, S.; Stiewe, T.; Brune, B.; et al. Reprogramming of tumor-associated macrophages by targeting beta-catenin/FOSL2/ARID5A signaling: A potential treatment of lung cancer. Sci. Adv. 2020, 6, eaaz6105. [Google Scholar] [CrossRef]
- Renoux, F.; Stellato, M.; Haftmann, C.; Vogetseder, A.; Huang, R.; Subramaniam, A.; Becker, M.O.; Blyszczuk, P.; Becher, B.; Distler, J.H.W.; et al. The AP1 Transcription Factor Fosl2 Promotes Systemic Autoimmunity and Inflammation by Repressing Treg Development. Cell Rep. 2020, 31, 107826. [Google Scholar] [CrossRef]
- Zomer, H.D.; Trentin, A.G. Skin wound healing in humans and mice: Challenges in translational research. J. Dermatol. Sci. 2018, 90, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Mirza, R.; Kwon, Y.; DiPietro, L.A.; Koh, T.J. The murine excisional wound model: Contraction revisited. Wound Repair Regen. 2015, 23, 874–877. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ge, J.; Tredget, E.E.; Wu, Y. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat. Protoc. 2013, 8, 302–309. [Google Scholar] [CrossRef]
- Gorell, E.; Nguyen, N.; Lane, A.; Siprashvili, Z. Gene therapy for skin diseases. Cold Spring Harb. Perspect. Med. 2014, 4, a015149. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Mei, S.; Dong, Y.; She, F.; Li, Y.; Li, P.; Kong, L. Functional Nanofibrous Biomaterials of Tailored Structures for Drug Delivery-A Critical Review. Pharmaceutics 2020, 12, 522. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhattacharya, N.; Indra, A.K.; Ganguli-Indra, G. Selective Ablation of BCL11A in Epidermal Keratinocytes Alters Skin Homeostasis and Accelerates Excisional Wound Healing In Vivo. Cells 2022, 11, 2106. https://doi.org/10.3390/cells11132106
Bhattacharya N, Indra AK, Ganguli-Indra G. Selective Ablation of BCL11A in Epidermal Keratinocytes Alters Skin Homeostasis and Accelerates Excisional Wound Healing In Vivo. Cells. 2022; 11(13):2106. https://doi.org/10.3390/cells11132106
Chicago/Turabian StyleBhattacharya, Nilika, Arup K. Indra, and Gitali Ganguli-Indra. 2022. "Selective Ablation of BCL11A in Epidermal Keratinocytes Alters Skin Homeostasis and Accelerates Excisional Wound Healing In Vivo" Cells 11, no. 13: 2106. https://doi.org/10.3390/cells11132106
APA StyleBhattacharya, N., Indra, A. K., & Ganguli-Indra, G. (2022). Selective Ablation of BCL11A in Epidermal Keratinocytes Alters Skin Homeostasis and Accelerates Excisional Wound Healing In Vivo. Cells, 11(13), 2106. https://doi.org/10.3390/cells11132106