A Resilience Related Glial-Neurovascular Network Is Transcriptionally Activated after Chronic Social Defeat in Male Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Chronic Social Defeat Stress
2.3. Social Interaction Test
2.4. Modified Social Interaction Test
2.5. Drug Administration
2.6. Tissue Dissection and Dissociation
2.7. Cell Collection
2.8. Single-Cell Sequencing
2.9. Bioinformatic Processing
2.10. Predicted Ligand-Receptor Interaction Analysis
2.11. Statistical Analysis of Behavioural Data
3. Results
3.1. Characterisation of Distinct Behavioural Outcomes after CSDS
3.2. Single-Cell RNA-Seq Analysis Identifies 41 Distinct Cell Clusters in the Hippocampus
3.3. Specific Cell-Type CSDS Responses in the Defeated Mice
3.4. The Microglia in the CSDS Response
3.5. The Oligodendrocytes in the CSDS Response
3.6. The BBB in the CSDS Response
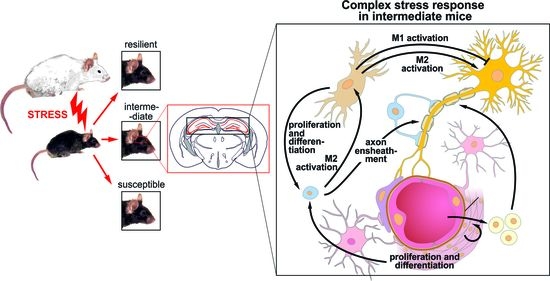
3.7. Cell-Cell Interaction after CSDS
3.8. Rapamycin Drug Treatment Improves Stress Resilience
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalisch, R.; Baker, D.G.; Basten, U.; Boks, M.P.; Bonanno, G.A.; Brummelman, E.; Chmitorz, A.; Fernàndez, G.; Fiebach, C.J.; Galatzer-Levy, I.; et al. The Resilience Framework as a Strategy to Combat Stress-Related Disorders. Nat. Hum. Behav. 2017, 1, 784–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalisch, R.; Cramer, A.O.J.; Binder, H.; Fritz, J.; Leertouwer, I.J.; Lunansky, G.; Meyer, B.; Timmer, J.; Veer, I.M.; van Harmelen, A.L. Deconstructing and Reconstructing Resilience: A Dynamic Network Approach. Perspect. Psychol. Sci. 2019, 14, 765–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, S.R.; Feder, A. Understanding Resilience and Preventing and Treating PTSD. Harv. Rev. Psychiatry 2018, 26, 158–174. [Google Scholar] [CrossRef] [PubMed]
- Rakesh, G.; Morey, R.A.; Zannas, A.S.; Malik, Z.; Marx, C.E.; Clausen, A.N.; Kritzer, M.D.; Szabo, S.T. Resilience as a Translational Endpoint in the Treatment of PTSD. Mol. Psychiatry 2019, 24, 1268–1283. [Google Scholar] [CrossRef]
- Santomauro, D.F.; Mantilla Herrera, A.M.; Shadid, J.; Zheng, P.; Ashbaugh, C.; Pigott, D.M.; Abbafati, C.; Adolph, C.; Amlag, J.O.; Aravkin, A.Y.; et al. Global Prevalence and Burden of Depressive and Anxiety Disorders in 204 Countries and Territories in 2020 Due to the COVID-19 Pandemic. Lancet 2021, 398, 1700–1712. [Google Scholar] [CrossRef]
- Russo, S.J.; Murrough, J.W.; Han, M.H.; Charney, D.S.; Nestler, E.J. Neurobiology of Resilience. Nat. Neurosci. 2012, 15, 1475–1484. [Google Scholar] [CrossRef] [Green Version]
- Ménard, C.; Pfau, M.L.; Hodes, G.E.; Russo, S.J. Immune and Neuroendocrine Mechanisms of Stress Vulnerability and Resilience. Neuropsychopharmacology 2017, 42, 62–80. [Google Scholar] [CrossRef] [Green Version]
- Cathomas, F.; Murrough, J.W.; Nestler, E.J.; Han, M.H.; Russo, S.J. Neurobiology of Resilience: Interface Between Mind and Body. Biol. Psychiatry 2019, 86, 410–420. [Google Scholar] [CrossRef]
- Dudek, K.A.; Dion-Albert, L.; Kaufmann, F.N.; Tuck, E.; Lebel, M.; Menard, C. Neurobiology of Resilience in Depression: Immune and Vascular Insights from Human and Animal Studies. Eur. J. Neurosci. 2021, 53, 183–221. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, V.; Han, M.H.; Graham, D.L.; Berton, O.; Renthal, W.; Russo, S.J.; LaPlant, Q.; Graham, A.; Lutter, M.; Lagace, D.C.; et al. Molecular Adaptations Underlying Susceptibility and Resistance to Social Defeat in Brain Reward Regions. Cell 2007, 131, 391–404. [Google Scholar] [CrossRef]
- Bagot, R.C.C.; Cates, H.M.M.; Purushothaman, I.; Lorsch, Z.S.S.; Walker, D.M.M.; Wang, J.; Huang, X.; Schlüter, O.M.M.; Maze, I.; Peña, C.J.J.; et al. Circuit-Wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron 2016, 90, 969–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasca, C.; Bigio, B.; Zelli, D.; de Angelis, P.; Lau, T.; Okamoto, M.; Soya, H.; Ni, J.; Brichta, L.; Greengard, P.; et al. Role of the Astroglial Glutamate Exchanger XCT in Ventral Hippocampus in Resilience to Stress. Neuron 2017, 96, 402–413.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorsch, Z.S.; Loh, Y.H.E.; Purushothaman, I.; Walker, D.M.; Parise, E.M.; Salery, M.; Cahill, M.E.; Hodes, G.E.; Pfau, M.L.; Kronman, H.; et al. Estrogen Receptor α Drives Pro-Resilient Transcription in Mouse Models of Depression. Nat. Commun. 2018, 9, 1116. [Google Scholar] [CrossRef] [PubMed]
- Herculano-Houzel, S. The Glia/Neuron Ratio: How It Varies Uniformly across Brain Structures and Species and What That Means for Brain Physiology and Evolution. Glia 2014, 62, 1377–1391. [Google Scholar] [CrossRef]
- D’ambrosi, N.; Cozzolino, M.; Apolloni, S. The Contribution of Non-Neuronal Cells in Neurodegeneration: From Molecular Pathogenesis to Therapeutic Challenges. Cells 2022, 11, 193. [Google Scholar] [CrossRef]
- Crabé, R.; Aimond, F.; Gosset, P.; Scamps, F.; Raoul, C. How Degeneration of Cells Surrounding Motoneurons Contributes to Amyotrophic Lateral Sclerosis. Cells 2020, 9, 2550. [Google Scholar] [CrossRef]
- Menard, C.; Pfau, M.L.; Hodes, G.E.; Kana, V.; Wang, V.X.; Bouchard, S.; Takahashi, A.; Flanigan, M.E.; Aleyasin, H.; Leclair, K.B.; et al. Social Stress Induces Neurovascular Pathology Promoting Depression. Nat. Neurosci. 2017, 20, 1752–1760. [Google Scholar] [CrossRef] [Green Version]
- Laine, M.A.; Trontti, K.; Misiewicz, Z.; Sokolowska, E.; Kulesskaya, N.; Heikkinen, A.; Saarnio, S.; Balcells, I.; Ameslon, P.; Greco, D.; et al. Genetic Control of Myelin Plasticity after Chronic Psychosocial Stress. eNeuro 2018, 5, ENEURO.0166-18.2018. [Google Scholar] [CrossRef]
- Bonnefil, V.; Dietz, K.; Amatruda, M.; Wentling, M.; Aubry, A.V.; Dupree, J.L.; Temple, G.; Park, H.J.; Burghardt, N.S.; Casaccia, P.; et al. Region-Specific Myelin Differences Define Behavioral Consequences of Chronic Social Defeat Stress in Mice. Elife 2019, 8, e40855. [Google Scholar] [CrossRef]
- Demin, K.A.; Smagin, D.A.; Kovalenko, I.L.; Strekalova, T.; Galstyan, D.S.; Kolesnikova, T.O.; De Abreu, M.S.; Galyamina, A.G.; Bashirzade, A.; Kalueff, A.V. CNS Genomic Profiling in the Mouse Chronic Social Stress Model Implicates a Novel Category of Candidate Genes Integrating Affective Pathogenesis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 105, 110086. [Google Scholar] [CrossRef]
- Ayash, S.; Lingner, T.; Ryu, S.; Kalisch, R.; Schmitt, U.; Müller, M.B. Reconceptualising Resilience within a Translational Framework Is Supported by Unique and Brain-Region Specific Transcriptional Signatures in Mice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ayash, S.; Schmitt, U.; Müller, M.B. Chronic Social Defeat-Induced Social Avoidance as a Proxy of Stress Resilience in Mice Involves Conditioned Learning. J. Psychiatr. Res. 2020, 120, 64–71. [Google Scholar] [CrossRef]
- Wendelmuth, M.; Willam, M.; Todorov, H.; Radyushkin, K.; Gerber, S.; Schweiger, S. Dynamic Longitudinal Behavior in Animals Exposed to Chronic Social Defeat Stress. PLoS ONE 2020, 15, e0235268. [Google Scholar] [CrossRef]
- Bonanno, G.A. Loss, Trauma, and Human Resilience: Have We Underestimated the Human Capacity to Thrive after Extremely Aversive Events? Am. Psychol. 2004, 59, 20–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonanno, G.A.; Westphal, M.; Mancini, A.D. Resilience to Loss and Potential Trauma. Annu. Rev. Clin. Psychol. 2011, 7, 511–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golden, S.A.; Covington, H.E., III; Berton, O.; Russo, S.J. A Standardized Protocol for Repeated Social Defeat Stress in Mice. Nat. Protoc. 2011, 6, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Van Der Kooij, M.A.; Jene, T.; Treccani, G.; Miederer, I.; Hasch, A.; Voelxen, N.; Walenta, S.; Müller, M.B. Chronic Social Stress-Induced Hyperglycemia in Mice Couples Individual Stress Susceptibility to Impaired Spatial Memory. Proc. Natl. Acad. Sci. USA 2018, 115, E10187–E10196. [Google Scholar] [CrossRef] [Green Version]
- Campbell, J.N.; Macosko, E.Z.; Fenselau, H.; Pers, T.H.; Lyubetskaya, A.; Tenen, D.; Goldman, M.; Verstegen, A.M.J.; Resch, J.M.; McCarroll, S.A.; et al. A Molecular Census of Arcuate Hypothalamus and Median Eminence Cell Types. Nat. Neurosci. 2017, 20, 484–496. [Google Scholar] [CrossRef] [Green Version]
- Gerber, S.; Pospisil, L.; Navandar, M.; Horenko, I. Low-Cost Scalable Discretization, Prediction, and Feature Selection for Complex Systems. Sci. Adv. 2020, 6, eaaw0961. [Google Scholar] [CrossRef] [Green Version]
- Horenko, I. On a Scalable Entropic Breaching of the Overfitting Barrier for Small Data Problems in Machine Learning. Neural Comput. 2020, 32, 1563–1579. [Google Scholar] [CrossRef]
- Shao, X.; Liao, J.; Li, C.; Lu, X.; Cheng, J.; Fan, X. CellTalkDB: A Manually Curated Database of Ligand–Receptor Interactions in Humans and Mice. Brief. Bioinform. 2020, 22, bbaa269. [Google Scholar] [CrossRef] [PubMed]
- Higashida, S.; Nagai, H.; Nakayama, K.; Shinohara, R.; Taniguchi, M.; Nagai, M.; Hikida, T.; Yawata, S.; Ago, Y.; Kitaoka, S.; et al. Repeated social defeat stress impairs attentional set shifting irrespective of social avoidance and increases female preference associated with heightened anxiety. Sci. Rep. 2018, 8, 10454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šabanović, M.; Liu, H.; Mlambo, V.; Aqel, H.; Chaudhury, D. What it takes to be at the top: The interrelationship between chronic social stress and social dominance. Brain Behav. 2020, 10, e01896. [Google Scholar] [CrossRef] [PubMed]
- Morel, C.; Montgomery, S.E.; Li, L.; Durand-de Cuttoli, R.; Teichman, E.M.; Juarez, B.; Tzavaras, N.; Ku, S.M.; Flanigan, M.E.; Cai, M.; et al. Midbrain projection to the basolateral amygdala encodes anxiety-like but not depression-like behaviors. Nat. Commun. 2022, 13, 1532. [Google Scholar] [CrossRef]
- Ximerakis, M.; Lipnick, S.L.; Innes, B.T.; Simmons, S.K.; Adiconis, X.; Dionne, D.; Mayweather, B.A.; Nguyen, L.; Niziolek, Z.; Ozek, C.; et al. Single-Cell Transcriptomic Profiling of the Aging Mouse Brain. Nat. Neurosci. 2019, 22, 1696–1708. [Google Scholar] [CrossRef]
- Bakulski, K.M.; Dou, J.F.; Thompson, R.C.; Lee, C.; Middleton, L.Y.; Perera, B.P.U.; Ferris, S.P.; Jones, T.R.; Neier, K.; Zhou, X.; et al. Single-Cell Analysis of the Gene Expression Effects of Developmental Lead (Pb) Exposure on the Mouse Hippocampus. Toxicol. Sci. 2020, 176, 396–409. [Google Scholar] [CrossRef]
- Bouvier, E.; Brouillard, F.; Molet, J.; Claverie, D.; Cabungcal, J.H.; Cresto, N.; Doligez, N.; Rivat, C.; Do, K.Q.; Bernard, C.; et al. Nrf2-dependent persistent oxidative stress results in stress-induced vulnerability to depression. Mol. Psychiatry 2016, 22, 1701–1713. [Google Scholar] [CrossRef] [Green Version]
- Stankiewicz, A.M.; Goscik, J.; Swiergiel, A.H.; Majewska, A.; Wieczorek, M.; Juszczak, G.R.; Lisowski, P. Social Stress Increases Expression of Hemoglobin Genes in Mouse Prefrontal Cortex. BMC Neurosci. 2014, 15, 130. [Google Scholar] [CrossRef] [Green Version]
- Wohleb, E.S.; Franklin, T.; Iwata, M.; Duman, R.S. Integrating Neuroimmune Systems in the Neurobiology of Depression. Nat. Rev. Neurosci. 2016, 17, 497–511. [Google Scholar] [CrossRef]
- Subramaniam, S.R.; Federoff, H.J. Targeting Microglial Activation States as a Therapeutic Avenue in Parkinson’s Disease. Front. Aging Neurosci. 2017, 9, 176. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, S.; Gritti, L.; Crooks, D.; Dombrowski, Y. Oligodendrocytes in Development, Myelin Generation and Beyond. Cells 2019, 8, 1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, B.; Zhu, Z.; Ransom, B.R.; Tong, X. Oligodendrocyte Lineage Cells and Depression. Mol. Psychiatry 2020, 26, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Peferoen, L.; Kipp, M.; van der Valk, P.; van Noort, J.M.; Amor, S. Oligodendrocyte-Microglia Cross-Talk in the Central Nervous System. Immunology 2014, 141, 302–313. [Google Scholar] [CrossRef]
- Bohlson, S.S.; O’Conner, S.D.; Hulsebus, H.J.; Ho, M.M.; Fraser, D.A. Complement, C1Q, and C1q-Related Molecules Regulate Macrophage Polarization. Front. Immunol. 2014, 5, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traiffort, E.; Kassoussi, A.; Zahaf, A.; Laouarem, Y. Astrocytes and Microglia as Major Players of Myelin Production in Normal and Pathological Conditions. Front. Cell. Neurosci. 2020, 14, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, T.; Sankowski, R.; Staszewski, O.; Prinz, M. Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 2020, 30, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Lécuyer, M.A.; Kebir, H.; Prat, A. Glial Influences on BBB Functions and Molecular Players in Immune Cell Trafficking. Biochim. Biophys. Acta-Mol. Basis Dis. 2016, 1862, 472–482. [Google Scholar] [CrossRef]
- Parker, K.R.; Migliorini, D.; Perkey, E.; Yost, K.E.; Bhaduri, A.; Bagga, P.; Haris, M.; Wilson, N.E.; Liu, F.; Gabunia, K.; et al. Single-Cell Analyses Identify Brain Mural Cells Expressing CD19 as Potential Off-Tumor Targets for CAR-T Immunotherapies. Cell 2020, 183, 126–142.e17. [Google Scholar] [CrossRef]
- Duan, L.; Zhang, X.D.; Miao, W.Y.; Sun, Y.J.; Xiong, G.; Wu, Q.; Li, G.; Yang, P.; Yu, H.; Li, H.; et al. PDGFRβ Cells Rapidly Relay Inflammatory Signal from the Circulatory System to Neurons via Chemokine CCL2. Neuron 2018, 100, 183–200.e8. [Google Scholar] [CrossRef] [Green Version]
- Moon, B.S.; Yoon, J.Y.; Kim, M.Y.; Lee, S.H.; Choi, T.; Choi, K.Y. Bone Morphogenetic Protein 4 Stimulates Neuronal Differentiation of Neuronal Stem Cells through the ERK Pathway. Exp. Mol. Med. 2009, 41, 116–125. [Google Scholar] [CrossRef]
- El-Ghissassi, F.; Valsesia-Wittmann, S.; Falette, N.; Duriez, C.; Walden, P.D.; Puisieux, A. BTG2TIS21/PC3 Induces Neuronal Differentiation and Prevents Apoptosis of Terminally Differentiated PC12 Cells. Oncogene 2002, 21, 6772–6778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurwitz, D.; Cunningham, D.D. Neurite Outgrowth Activity of Protease Nexin-1 on Neuroblastoma Cells Requires Thrombin Inhibition. J. Cell. Physiol. 1990, 142, 155–162. [Google Scholar] [CrossRef]
- De La Fuente, A.G.; Lange, S.; Silva, M.E.; Gonzalez, G.A.; Tempfer, H.; van Wijngaarden, P.; Zhao, C.; Di Canio, L.; Trost, A.; Bieler, L.; et al. Pericytes Stimulate Oligodendrocyte Progenitor Cell Differentiation during CNS Remyelination. Cell Rep. 2017, 20, 1755–1764. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, N.; Terlizzi, J.; Aho, S.; Brittingham, R.; Fertala, A.; Oyama, N.; McGrath, J.A.; Uitto, J. Extracellular Matrix Protein 1 Inhibits the Activity of Matrix Metalloproteinase 9 through High-Affinity Protein/Protein Interactions. Exp. Dermatol. 2006, 15, 300–307. [Google Scholar] [CrossRef]
- Osman, B.; Akool, E.S.; Doller, A.; Müller, R.; Pfeilschifter, J.; Eberhardt, W. Differential Modulation of the Cytokine-Induced MMP-9/TIMP-1 Protease-Antiprotease System by the MTOR Inhibitor Rapamycin. Biochem. Pharmacol. 2011, 81, 134–143. [Google Scholar] [CrossRef] [Green Version]
- Stamatovic, S.M.; Johnson, A.M.; Keep, R.F.; Andjelkovic, A.V. Junctional Proteins of the Blood-Brain Barrier: New Insights into Function and Dysfunction. Tissue Barriers 2016, 4, e1154641. [Google Scholar] [CrossRef] [Green Version]
- Baeten, K.M.; Akassoglou, K. Extracellular Matrix and Matrix Receptors in Blood-Brain Barrier Formation and Stroke. Dev. Neurobiol. 2011, 71, 1018–1039. [Google Scholar] [CrossRef] [Green Version]
- Bockaert, J.; Marin, P. MTOR in Brain Physiology and Pathologies. Physiol. Rev. 2015, 95, 1157–1187. [Google Scholar] [CrossRef] [Green Version]
- You, W.; Wang, Z.; Li, H.; Shen, H.; Xu, X.; Jia, G.; Chen, G. Inhibition of Mammalian Target of Rapamycin Attenuates Early Brain Injury through Modulating Microglial Polarization after Experimental Subarachnoid Hemorrhage in Rats. J. Neurol. Sci. 2016, 367, 224–231. [Google Scholar] [CrossRef]
- Xu, X.; Gao, W.; Li, L.; Hao, J.; Yang, B.; Wang, T.; Li, L.; Bai, X.; Li, F.; Ren, H.; et al. Annexin A1 Protects against Cerebral Ischemia–Reperfusion Injury by Modulating Microglia/Macrophage Polarization via FPR2/ALX-Dependent AMPK-MTOR Pathway. J. Neuroinflammation 2021, 18, 119. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Li, D.; Zhang, N.; Liu, R.; Han, B.; Wei, C.; Liu, H.; Xu, X.; Hao, J. Everolimus (RAD001) Ameliorates Vascular Cognitive Impairment by Regulating Microglial Function via the MTORC1 Signaling Pathway. J. Neuroimmunol. 2016, 299, 164–171. [Google Scholar] [CrossRef]
- Harry, G.J. Microglia during Development and Aging. Pharmacol. Ther. 2013, 139, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Van Vliet, E.A.; Otte, W.M.; Wadman, W.J.; Aronica, E.; Kooij, G.; De Vries, H.E.; Dijkhuizen, R.M.; Gorter, J.A. Blood-Brain Barrier Leakage after Status Epilepticus in Rapamycin-Treated Rats II: Potential Mechanisms. Epilepsia 2016, 57, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Chi, O.Z.; Kiss, G.K.; Mellender, S.J.; Liu, X.; Weiss, H.R. Rapamycin Decreased Blood-Brain Barrier Permeability in Control but Not in Diabetic Rats in Early Cerebral Ischemia. Neurosci. Lett. 2017, 654, 17–22. [Google Scholar] [CrossRef]
- Van Skike, C.E.; Lin, A.L.; Roberts Burbank, R.; Halloran, J.J.; Hernandez, S.F.; Cuvillier, J.; Soto, V.Y.; Hussong, S.A.; Jahrling, J.B.; Javors, M.A.; et al. MTOR Drives Cerebrovascular, Synaptic, and Cognitive Dysfunction in Normative Aging. Aging Cell 2020, 19, e13057. [Google Scholar] [CrossRef]
- Albert, L.; Karsy, M.; Murali, R.; Jhanwar-Uniyal, M. Inhibition of mTOR Activates the MAPK Pathway in Glioblastoma Multiforme. Cancer Genom. Proteom. 2009, 6, 255–261. [Google Scholar]
- Stein, D.J.; Vasconcelos, M.F.; Albrechet-Souza, L.; Ceresér, K.M.M.; De Almeida, R.M.M. Microglial Over-Activation by Social Defeat Stress Contributes to Anxiety-and Depressive-like Behaviors. Front. Behav. Neurosci. 2017, 11, 207. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.R.; Liu, J.C.; Bao, J.S.; Bai, Q.Q.; Wang, G.Q. Interaction of Microglia and Astrocytes in the Neurovascular Unit. Front. Immunol. 2020, 11, 1024. [Google Scholar] [CrossRef]
- Lehmann, M.L.; Weigel, T.K.; Elkahloun, A.G.; Herkenham, M. Chronic Social Defeat Reduces Myelination in the Mouse Medial Prefrontal Cortex. Sci. Rep. 2017, 7, 46548. [Google Scholar] [CrossRef] [Green Version]
- Van Noort, J.M.; Bsibsi, M.; Gerritsen, W.H.; Van Der Valk, P.; Bajramovic, J.J.; Steinman, L.; Amor, S. AB-Crystallin Is a Target for Adaptive Immune Responses and a Trigger of Innate Responses in Preactive Multiple Sclerosis Lesions. J. Neuropathol. Exp. Neurol. 2010, 69, 694–703. [Google Scholar] [CrossRef] [Green Version]
- Shen, K.; Reichelt, M.; Kyauk, R.V.; Ngu, H.; Shen, Y.A.A.; Foreman, O.; Modrusan, Z.; Friedman, B.A.; Sheng, M.; Yuen, T.J. Multiple Sclerosis Risk Gene Mertk Is Required for Microglial Activation and Subsequent Remyelination. Cell Rep. 2021, 34, 108835. [Google Scholar] [CrossRef]
- Han, Y.; Sun, C.Y.; Meng, S.Q.; Tabarak, S.; Yuan, K.; Cao, L.; Yan, W.; Xu, L.Z.; Deng, J.H.; Zhu, W.L.; et al. Systemic Immunization with Altered Myelin Basic Protein Peptide Produces Sustained Antidepressant-like Effects. Mol. Psychiatry 2020, 25, 1260–1274. [Google Scholar] [CrossRef]
- Czéh, B.; Varga, Z.K.K.; Henningsen, K.; Kovács, G.L.; Miseta, A.; Wiborg, O. Chronic Stress Reduces the Number of GABAergic Interneurons in the Adult Rat Hippocampus, Dorsal-Ventral and Region-Specific Differences. Hippocampus 2015, 25, 393–405. [Google Scholar] [CrossRef]
- Pinto, V.; Costa, J.C.; Morgado, P.; Mota, C.; Miranda, A.; Bravo, F.V.; Oliveira, T.G.; Cerqueira, J.J.; Sousa, N. Differential Impact of Chronic Stress along the Hippocampal Dorsal–Ventral Axis. Brain Struct. Funct. 2015, 220, 1205–1212. [Google Scholar] [CrossRef] [Green Version]
- MacCallum, P.E.; Blundell, J. The MTORC1 Inhibitor Rapamycin and the MTORC1/2 Inhibitor AZD2014 Impair the Consolidation and Persistence of Contextual Fear Memory. Psychopharmacology 2020, 237, 2795–2808. [Google Scholar] [CrossRef]
- Lopatynska-mazurek, M.; Komsta, L.; Gibula-tarlowska, E.; Kotlinska, J.H. Aversive Learning Deficits and Depressive-Like Behaviors Are Accompanied by an Increase in Oxidative Stress in a Rat Model of Fetal Alcohol Spectrum Disorders: The Protective Effect of Rapamycin. Int. J. Mol. Sci. 2021, 22, 7083. [Google Scholar] [CrossRef]
- Gafford, G.M.; Parsons, R.G.; Helmstetter, F.J. Consolidation and Reconsolidation of Contextual Fear Memory Requires Mammalian Target of Rapamycin-Dependent Translation in the Dorsal Hippocampus. Neuroscience 2011, 182, 98–104. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vennin, C.; Hewel, C.; Todorov, H.; Wendelmuth, M.; Radyushkin, K.; Heimbach, A.; Horenko, I.; Ayash, S.; Müller, M.B.; Schweiger, S.; et al. A Resilience Related Glial-Neurovascular Network Is Transcriptionally Activated after Chronic Social Defeat in Male Mice. Cells 2022, 11, 3405. https://doi.org/10.3390/cells11213405
Vennin C, Hewel C, Todorov H, Wendelmuth M, Radyushkin K, Heimbach A, Horenko I, Ayash S, Müller MB, Schweiger S, et al. A Resilience Related Glial-Neurovascular Network Is Transcriptionally Activated after Chronic Social Defeat in Male Mice. Cells. 2022; 11(21):3405. https://doi.org/10.3390/cells11213405
Chicago/Turabian StyleVennin, Constance, Charlotte Hewel, Hristo Todorov, Marlon Wendelmuth, Konstantin Radyushkin, André Heimbach, Illia Horenko, Sarah Ayash, Marianne B. Müller, Susann Schweiger, and et al. 2022. "A Resilience Related Glial-Neurovascular Network Is Transcriptionally Activated after Chronic Social Defeat in Male Mice" Cells 11, no. 21: 3405. https://doi.org/10.3390/cells11213405
APA StyleVennin, C., Hewel, C., Todorov, H., Wendelmuth, M., Radyushkin, K., Heimbach, A., Horenko, I., Ayash, S., Müller, M. B., Schweiger, S., Gerber, S., & Lutz, B. (2022). A Resilience Related Glial-Neurovascular Network Is Transcriptionally Activated after Chronic Social Defeat in Male Mice. Cells, 11(21), 3405. https://doi.org/10.3390/cells11213405