Three-dimensional Organization of Polytene Chromosomes in Somatic and Germline Tissues of Malaria Mosquitoes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mosquito Species and Strains
2.2. Isolation of Tissues
2.3. Design of Oligonucleotide Probes
2.4. Labeling of Oligonucleotides
2.5. FISH with Oligopaints
2.6. Whole-Mount Immunostaining
2.7. Confocal Microscopy
2.8. Processing and Analysis of Images
2.9. Statistical Analyses of Data
3. Results
3.1. Development of A Workflow to Characterize Oligopainted Chromosome Territories in Anopheles
3.2. The Volume and Shape of Chromosome Territories in Anopheles
3.3. Polarization of Chromosome Territories Indicate A Rabl-like Configuration in Germline and Soma
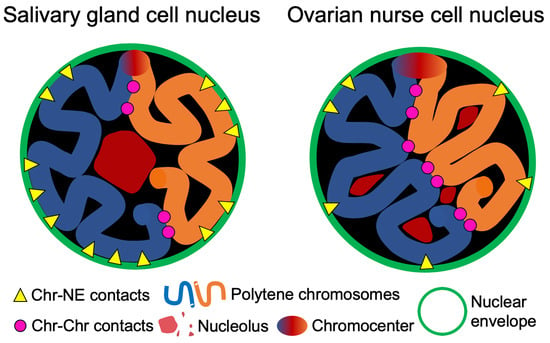
3.4. Contacts of Chromosome Territories with the Nuclear Envelope in Malaria Mosquitoes
3.5. Chromosome–Chromosome Contacts in Malaria Mosquitoes
3.6. An Inverse Relationship between Chromosome–Nuclear Envelope and Chromosome–Chromosome Contacts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cremer, T.; Kreth, G.; Koester, H.; Fink, R.H.A.; Heintzmann, R.; Cremer, M.; Solovei, I.; Zink, D.; Cremer, C. Chromosome territories, interchromatin domain compartment, and nuclear matrix: An integrated view of the functional nuclear architecture. Crit. Rev. Eukar. Gene 2000, 10, 179–212. [Google Scholar] [CrossRef] [Green Version]
- Lanctot, C.; Cheutin, T.; Cremer, M.; Cavalli, G.; Cremer, T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007, 8, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Sexton, T.; Cavalli, G. The role of chromosome domains in shaping the functional genome. Cell 2015, 160, 1049–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharakhov, I.V.; Bondarenko, S.M.; Artemov, G.N.; Onufriev, A.V. The Role of Chromosome-Nuclear Envelope Attachments in 3D Genome Organization. Biochem. (Mosc.) 2018, 83, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Van Steensel, B.; Furlong, E.E.M. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M. Chromosome Territories. Csh Perspect. Biol. 2010, 2. [Google Scholar] [CrossRef] [Green Version]
- Fields, B.D.; Nguyen, S.C.; Nir, G.; Kennedy, S. A multiplexed DNA FISH strategy for assessing genome architecture in Caenorhabditis elegans. Elife 2019, 8. [Google Scholar] [CrossRef]
- Sexton, T.; Yaffe, E.; Kenigsberg, E.; Bantignies, F.; Leblanc, B.; Hoichman, M.; Parrinello, H.; Tanay, A.; Cavalli, G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012, 148, 458–472. [Google Scholar] [CrossRef] [Green Version]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Tanabe, H.; Muller, S.; Neusser, M.; von Hase, J.; Calcagno, E.; Cremer, M.; Solovei, I.; Cremer, C.; Cremer, T. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA 2002, 99, 4424–4429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mora, L.; Sanchez, I.; Garcia, M.; Ponsa, M. Chromosome territory positioning of conserved homologous chromosomes in different primate species. Chromosoma 2006, 115, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.; Fuchs, C.; Gonik, M.; Khrameeva, E.E.; Mironov, A.A.; Frishman, D. Inter-Chromosomal Contact Networks Provide Insights into Mammalian Chromatin Organization. PLoS ONE 2015, 10, e0126125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.A.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269. [Google Scholar] [CrossRef]
- Schubert, V.; Berr, A.; Meister, A. Interphase chromatin organisation in Arabidopsis nuclei: Constraints versus randomness. Chromosoma 2012, 121, 369–387. [Google Scholar] [CrossRef]
- Hochstrasser, M.; Mathog, D.; Gruenbaum, Y.; Saumweber, H.; Sedat, J.W. Spatial-Organization of Chromosomes in the Salivary-Gland Nuclei of Drosophila-Melanogaster. J. Cell Biol. 1986, 102, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Beliveau, B.J.; Apostolopoulos, N.; Wu, C.T. Visualizing genomes with Oligopaint FISH probes. Curr. Protoc. Mol. Biol. 2014, 105, Unit 14 23. [Google Scholar] [CrossRef]
- George, P.; Sharma, A.; Sharakhov, I.V. 2D and 3D chromosome painting in malaria mosquitoes. J. Vis. Exp. 2014, e51173. [Google Scholar] [CrossRef] [Green Version]
- Bondarenko, S.M.; Artemov, G.N.; Sharakhov, I.V.; Stegniy, V.N. Tissue-specific features of the X chromosome and nucleolus spatial dynamics in a malaria mosquito, Anopheles atroparvus. PLoS ONE 2017, 12, e0171290. [Google Scholar] [CrossRef]
- Erceg, J.; AlHaj Abed, J.; Goloborodko, A.; Lajoie, B.R.; Fudenberg, G.; Abdennur, N.; Imakaev, M.; McCole, R.B.; Nguyen, S.C.; Saylor, W.; et al. The genome-wide multi-layered architecture of chromosome pairing in early Drosophila embryos. Nat. Commun. 2019, 10, 4486. [Google Scholar] [CrossRef] [Green Version]
- Lowenstein, M.G.; Goddard, T.D.; Sedat, J.W. Long-range interphase chromosome organization in Drosophila: A study using color barcoded fluorescence in situ hybridization and structural clustering analysis. Mol. Biol. Cell 2004, 15, 5678–5692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branco, M.R.; Pombo, A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006, 4, e138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirny, L.A. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011, 19, 37–51. [Google Scholar] [CrossRef] [Green Version]
- Langersafer, P.R.; Levine, M.; Ward, D.C. Immunological Method for Mapping Genes on Drosophila Polytene Chromosomes. Proc. Natl. Acad. Sci. USA 1982, 79, 4381–4385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Steensel, B.; Dekker, J. Genomics tools for unraveling chromosome architecture. Nat. Biotechnol. 2010, 28, 1089–1095. [Google Scholar] [CrossRef] [Green Version]
- Belyaeva, A.; Venkatachalapathy, S.; Nagarajan, M.; Shivashankar, G.V.; Uhler, C. Network analysis identifies chromosome intermingling regions as regulatory hotspots for transcription. Proc. Natl. Acad. Sci. USA 2017, 114, 13714–13719. [Google Scholar] [CrossRef] [Green Version]
- Maharana, S.; Iyer, K.V.; Jain, N.; Nagarajan, M.; Wang, Y.; Shivashankar, G.V. Chromosome intermingling-the physical basis of chromosome organization in differentiated cells. Nucleic Acids Res. 2016, 44, 5148–5160. [Google Scholar] [CrossRef] [Green Version]
- Hochstrasser, M.; Sedat, J.W. Three-Dimensional Organization of Drosophila melanogaster Interphase Nuclei. 1. Tissue-Specific Aspects of Polytene Nuclear Architecture. J. Cell Biol. 1987, 104, 1455–1470. [Google Scholar] [CrossRef]
- Hochstrasser, M.; Sedat, J.W. Three-Dimensional Organization of Drosophila melanogaster Interphase Nuclei. 2. Chromosome Spatial Organization and Gene Regulation. J. Cell Biol. 1987, 104, 1471–1483. [Google Scholar] [CrossRef]
- Mathog, D.; Hochstrasser, M.; Gruenbaum, Y.; Saumweber, H.; Sedat, J. Characteristic Folding Pattern of Polytene Chromosomes in Drosophila Salivary-Gland Nuclei. Nature 1984, 308, 414–421. [Google Scholar] [CrossRef]
- Stegnii, V.N. Systemic reorganization of the architectonics of polytene chromosomes in the onto- and phylogenesis of malarial mosquitoes. II. Species specificity in the pattern of chromosome relations with the nuclear envelope of nutrient ovarian cells. Genetika 1987, 23, 1194–1199. [Google Scholar] [PubMed]
- Stegnii, V.N. Reorganization of the structure of interphase nuclei during the on to- and phylogeny of malaria mosquitoes. Dokl Akad Nauk Sssr 1979, 249, 1231–1234. [Google Scholar] [PubMed]
- Stegnii, V.N.; Sharakhova, M.V. Systemic reorganization of the architechtonics of polytene chromosomes in onto- and phylogenesis of malaria mosquitoes. Structural features regional of chromosomal adhesion to the nuclear membrane. Genetika 1991, 27, 828–835. [Google Scholar] [PubMed]
- Stegnii, V.N. Systemic reorganization of the architectonics of polytene chromosomes in the onto- and phylogenesis of malaria mosquitoes. Genetika 1987, 23, 821–827. [Google Scholar] [PubMed]
- Sharakhov, I.V.; Sharakhova, M.V.; Mbogo, C.M.; Koekemoer, L.L.; Yan, G. Linear and spatial organization of polytene chromosomes of the African malaria mosquito Anopheles funestus. Genetics 2001, 159, 211–218. [Google Scholar]
- Erenpreisa, J. Chromatin Organization in Interphase Cell Nucleus; Zinatne Publ: Riga, Latvia, 1990. [Google Scholar]
- Van Bemmel, J.G.; Pagie, L.; Braunschweig, U.; Brugman, W.; Meuleman, W.; Kerkhoven, R.M.; van Steensel, B. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE 2010, 5, e15013. [Google Scholar] [CrossRef] [Green Version]
- Pindyurin, A.V.; Ilyin, A.A.; Ivankin, A.V.; Tselebrovsky, M.V.; Nenasheva, V.V.; Mikhaleva, E.A.; Pagie, L.; van Steensel, B.; Shevelyov, Y.Y. The large fraction of heterochromatin in Drosophila neurons is bound by both B-type lamin and HP1a. Epigenetics Chromatin 2018, 11, 65. [Google Scholar] [CrossRef] [Green Version]
- Van Steensel, B.; Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000, 18, 424–428. [Google Scholar] [CrossRef]
- Bickmore, W.A.; van Steensel, B. Genome architecture: Domain organization of interphase chromosomes. Cell 2013, 152, 1270–1284. [Google Scholar] [CrossRef] [Green Version]
- Cavalli, G.; Misteli, T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 2013, 20, 290–299. [Google Scholar] [CrossRef]
- Chubb, J.R.; Boyle, S.; Perry, P.; Bickmore, W.A. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 2002, 12, 439–445. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Xu, X.; Nguyen, C.M.; Liu, Y.; Gao, Y.; Lin, X.; Daley, T.; Kipniss, N.H.; La Russa, M.; Qi, L.S. CRISPR-Mediated Programmable 3D Genome Positioning and Nuclear Organization. Cell 2018, 175, 1405–1417 e1414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, A.; Gasser, S.M. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007, 8, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, H.; Kalverda, B.; de Wit, E.; Talhout, W.; Fornerod, M.; van Steensel, B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006, 38, 1005–1014. [Google Scholar] [CrossRef]
- Kind, J.; Pagie, L.; Ortabozkoyun, H.; Boyle, S.; de Vries, S.S.; Janssen, H.; Amendola, M.; Nolen, L.D.; Bickmore, W.A.; van Steensel, B. Single-cell dynamics of genome-nuclear lamina interactions. Cell 2013, 153, 178–192. [Google Scholar] [CrossRef] [Green Version]
- Harr, J.C.; Luperchio, T.R.; Wong, X.R.; Cohen, E.; Wheelan, S.J.; Reddy, K.L. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J. Cell Biol. 2015, 208, 33–52. [Google Scholar] [CrossRef]
- Gonzalez-Sandoval, A.; Towbin, B.D.; Kalck, V.; Cabianca, D.S.; Gaidatzis, D.; Hauer, M.H.; Geng, L.Q.; Wang, L.; Yang, T.; Wang, X.H.; et al. Perinuclear Anchoring of H3K9-Methylated Chromatin Stabilizes Induced Cell Fate in C. elegans Embryos. Cell 2015, 163, 1333–1347. [Google Scholar] [CrossRef] [Green Version]
- Camps, J.; Erdos, M.R.; Ried, T. The role of lamin B1 for the maintenance of nuclear structure and function. Nucleus 2015, 6, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Kinney, N.A.; Sharakhov, I.V.; Onufriev, A.V. Chromosome-nuclear envelope attachments affect interphase chromosome territories and entanglement. Epigenetics Chromatin 2018, 11, 3. [Google Scholar] [CrossRef]
- Ulianov, S.V.; Doronin, S.A.; Khrameeva, E.E.; Kos, P.I.; Luzhin, A.V.; Starikov, S.S.; Galitsyna, A.A.; Nenasheva, V.V.; Ilyin, A.A.; Flyamer, I.M.; et al. Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 2019, 10, 1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinney, N.A.; Onufriev, A.V.; Sharakhov, I.V. Quantified effects of chromosome-nuclear envelope attachments on 3D organization of chromosomes. Nucleus 2015, 6, 212–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stormo, B.M.; Fox, D.T. Polyteny: Still a giant player in chromosome research. Chromosome Res. 2017, 25, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Zykova, T.Y.; Levitsky, V.G.; Belyaeva, E.S.; Zhimulev, I.F. Polytene Chromosomes—A Portrait of Functional Organization of the Drosophila Genome. Curr. Genom. 2018, 19, 179–191. [Google Scholar] [CrossRef]
- Eagen, K.P.; Hartl, T.A.; Kornberg, R.D. Stable Chromosome Condensation Revealed by Chromosome Conformation Capture. Cell 2015, 163, 934–946. [Google Scholar] [CrossRef] [Green Version]
- Kolesnikova, T.D.; Goncharov, F.P.; Zhimulev, I.F. Similarity in replication timing between polytene and diploid cells is associated with the organization of the Drosophila genome. PLoS ONE 2018, 13, e0195207. [Google Scholar] [CrossRef]
- Sharakhova, M.V.; George, P.; Brusentsova, I.V.; Leman, S.C.; Bailey, J.A.; Smith, C.D.; Sharakhov, I.V. Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genom. 2010, 11, 459. [Google Scholar] [CrossRef] [Green Version]
- Coluzzi, M.; Sabatini, A.; della Torre, A.; Di Deco, M.A.; Petrarca, V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 2002, 298, 1415–1418. [Google Scholar] [CrossRef]
- Coetzee, M.; Hunt, R.H.; Wilkerson, R.; Della Torre, A.; Coulibaly, M.B.; Besansky, N.J. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 2013, 3619, 246–274. [Google Scholar] [CrossRef] [Green Version]
- Barron, M.G.; Paupy, C.; Rahola, N.; Akone-Ella, O.; Ngangue, M.F.; Wilson-Bahun, T.A.; Pombi, M.; Kengne, P.; Costantini, C.; Simard, F.; et al. A new species in the major malaria vector complex sheds light on reticulated species evolution. Sci. Rep. 2019, 9, 14753. [Google Scholar] [CrossRef] [Green Version]
- Neafsey, D.E.; Waterhouse, R.M.; Abai, M.R.; Aganezov, S.S.; Alekseyev, M.A.; Allen, J.E.; Amon, J.; Arca, B.; Arensburger, P.; Artemov, G.; et al. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 2015, 347, 43. [Google Scholar] [CrossRef] [Green Version]
- Sharakhova, M.V.; Hammond, M.P.; Lobo, N.F.; Krzywinski, J.; Unger, M.F.; Hillenmeyer, M.E.; Bruggner, R.V.; Birney, E.; Collins, F.H. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 2007, 8, R5. [Google Scholar] [CrossRef] [Green Version]
- Holt, R.A.; Subramanian, G.M.; Halpern, A.; Sutton, G.G.; Charlab, R.; Nusskern, D.R.; Wincker, P.; Clark, A.G.; Ribeiro, J.M.; Wides, R.; et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science 2002, 298, 129–149. [Google Scholar] [CrossRef]
- George, P.; Sharakhova, M.V.; Sharakhov, I.V. High-resolution cytogenetic map for the African malaria vector Anopheles gambiae. Insect Mol. Biol. 2010, 19, 675–682. [Google Scholar] [CrossRef] [Green Version]
- Fontaine, M.C.; Pease, J.B.; Steele, A.; Waterhouse, R.M.; Neafsey, D.E.; Sharakhov, I.V.; Jiang, X.F.; Hall, A.B.; Catteruccia, F.; Kakani, E.; et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 2015, 347, 42. [Google Scholar] [CrossRef] [Green Version]
- Thawornwattana, Y.; Dalquen, D.; Yang, Z. Coalescent Analysis of Phylogenomic Data Confidently Resolves the Species Relationships in the Anopheles gambiae Species Complex. Mol. Biol. Evol. 2018, 35, 2512–2527. [Google Scholar] [CrossRef]
- Coluzzi, M.; Montalenti, S. Osservazioni comparative sul cromosoma X nelle specie A e B del comlesso Anopheles gambiae. Citologia 1966, 40, 671–678. [Google Scholar]
- Mal’ceva, N.I.; Gyurkovics, H.; Zhimulev, I.F. General characteristics of the polytene chromosome from ovarian pseudonurse cells of the Drosophila melanogaster otu11 and fs(2)B mutants. Chromosome Res. 1995, 3, 191–200. [Google Scholar] [CrossRef]
- King, R.C.; Riley, S.F.; Cassidy, J.D.; White, P.E.; Paik, Y.K. Giant polytene chromosomes from the ovaries of a Drosophila mutant. Science 1981, 212, 441–443. [Google Scholar] [CrossRef]
- Hartl, T.A.; Sweeney, S.J.; Knepler, P.J.; Bosco, G. Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genet. 2008, 4, e1000228. [Google Scholar] [CrossRef] [Green Version]
- Bauer, C.R.; Hartl, T.A.; Bosco, G. Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet. 2012, 8, e1002873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, J.A.; Brogdon, W.G.; Collins, F.H. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg. 1993, 49, 520–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fanello, C.; Santolamazza, F.; della Torre, A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med. Vet. Entomol. 2002, 16, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Rouillard, J.M.; Zuker, M.; Gulari, E. OligoArray 2.0: Design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003, 31, 3057–3062. [Google Scholar] [CrossRef]
- Murgha, Y.; Beliveau, B.; Semrau, K.; Schwartz, D.; Wu, C.T.; Gulari, E.; Rouillard, J.M. Combined in vitro transcription and reverse transcription to amplify and label complex synthetic oligonucleotide probe libraries. Biotechniques 2015, 58, 301–307. [Google Scholar] [CrossRef] [Green Version]
- Murgha, Y.E.; Rouillard, J.M.; Gulari, E. Methods for the preparation of large quantities of complex single-stranded oligonucleotide libraries. PLoS ONE 2014, 9, e94752. [Google Scholar] [CrossRef] [Green Version]
- MATLAB. MATLAB:2010 version 7.10.0 (R2010a); The MathWorks Inc.: Natick, MA, USA, 2010. [Google Scholar]
- Luong, B. Euclidian Projection on Ellipsoid and Conic. MATLAB Central File Exchange. Available online: https://www.mathworks.com/matlabcentral/fileexchange/27711-euclidian-projection-on-ellipsoid-and-conic (accessed on 22 January 2020).
- MATLAB. Scripts for Analyzing Chromosome Oligopaints. Figshare. Software. Available online: https://doi.org/10.6084/m9.figshare.11676312.v1 (accessed on 21 January 2020).
- Arcidiacono, G. Online Linear Regression Calculator. Available online: http://www.alcula.com/calculators/statistics/linear-regression/ (accessed on 17 January 2020).
- Grace 2D Graph Plotting Tool. Available online: http://plasma-gate.weizmann.ac.il/Grace/ (accessed on 17 January 2020).
- Stangroom, J. P Value from Pearson (R) Calculator. Available online: https://www.socscistatistics.com/pvalues/pearsondistribution.aspx (accessed on 17 January 2020).
- Jerabek, H.; Heermann, D.W. How Chromatin Looping and Nuclear Envelope Attachment Affect Genome Organization in Eukaryotic Cell Nuclei. Int. Rev. Cell. Mol. Biol. 2014, 307, 351–381. [Google Scholar] [CrossRef]
- Khalil, A.; Grant, J.L.; Caddle, L.B.; Atzema, E.; Mills, K.D.; Arneodo, A. Chromosome territories have a highly nonspherical morphology and nonrandom positioning. Chromosome Res. 2007, 15, 899–916. [Google Scholar] [CrossRef]
- Kinney, N.A.; Sharakhov, I.V.; Onufriev, A.V. Investigation of the chromosome regions with significant affinity for the nuclear envelope in fruit fly—A model based approach. PLoS ONE 2014, 9, e91943. [Google Scholar] [CrossRef]
- Khrameeva, E.E.; Fudenberg, G.; Gelfand, M.S.; Mirny, L.A. History of chromosome rearrangements reflects the spatial organization of yeast chromosomes. J. Bioinform. Comput. Biol. 2016, 14, 1641002. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Nagarajan, M.; Uhler, C.; Shivashankar, G.V. Orientation and repositioning of chromosomes correlate with cell geometry-dependent gene expression. Mol. Biol. Cell 2017, 28, 1997–2009. [Google Scholar] [CrossRef] [PubMed]
- Parada, L.A.; Misteli, T. Chromosome positioning in the interphase, nucleus. Trends Cell Biol. 2002, 12, 425–432. [Google Scholar] [CrossRef]
- Bystricky, K.; Laroche, T.; van Houwe, G.; Blaszczyk, M.; Gasser, S.M. Chromosome looping in yeast: Telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 2005, 168, 375–387. [Google Scholar] [CrossRef] [Green Version]
- Ostashevsky, J. A polymer model for large-scale chromatin organization in lower eukaryotes. Mol. Biol. Cell 2002, 13, 2157–2169. [Google Scholar] [CrossRef] [Green Version]
- Risca, V.I.; Greenleaf, W.J. Unraveling the 3D genome: Genomics tools for multiscale exploration. Trends Genet. Tig. 2015, 31, 357–372. [Google Scholar] [CrossRef] [Green Version]
- Knoch, T.A.; Munkel, C.; Langowski, J. Three-dimensional organization of chromosome territories and the human interphase cell nucleus-simulations versus experiments. J. Mol. Graph. Model. 1998, 16, 284. [Google Scholar] [CrossRef]
- Zink, D.; Cremer, T.; Saffrich, R.; Fischer, R.; Trendelenburg, M.F.; Ansorge, W.; Stelzer, E.H.K. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 1998, 102, 241–251. [Google Scholar] [CrossRef]
- Sadoni, N.; Langer, S.; Fauth, C.; Bernardi, G.; Cremer, T.; Turner, B.M.; Zink, D. Nuclear organization of mammalian genomes: Polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 1999, 146, 1211–1226. [Google Scholar] [CrossRef] [Green Version]
- Kreth, G.; Finsterle, J.; von Hase, J.; Cremer, M.; Cremer, C. Radial arrangement of chromosome territories in human cell nuclei: A computer model approach based on gene density indicates a probabilistic global positioning code. Biophys J. 2004, 86, 2803–2812. [Google Scholar] [CrossRef] [Green Version]
- Mora, L.; Sanchez, I.; Garcia, M.; Ponsa, M. Radial chromosome territories position analysis of five conserved chromosomes in three primate species. Chromosome Res. 2005, 13, 100–101. [Google Scholar]
- Artemov, G.N.; Bondarenko, S.M.; Naumenko, A.N.; Stegniy, V.N.; Sharakhova, M.V.; Sharakhov, I.V. Partial-arm translocations in evolution of malaria mosquitoes revealed by high-coverage physical mapping of the Anopheles atroparvus genome. BMC Genom. 2018, 19, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandhi, M.; Medvedovic, M.; Stringer, J.R.; Nikiforov, Y.E. Interphase chromosome folding determines spatial proximity of genes participating in carcinogenic RET/PTC rearrangements. Oncogene 2006, 25, 2360–2366. [Google Scholar] [CrossRef] [Green Version]
- Fudenberg, G.; Getz, G.; Meyerson-, M.; Mirny, L.A. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat. Biotechnol. 2011, 29, 1109–U1175. [Google Scholar] [CrossRef] [Green Version]
- Duan, Z.; Andronescu, M.; Schutz, K.; McIlwain, S.; Kim, Y.J.; Lee, C.; Shendure, J.; Fields, S.; Blau, C.A.; Noble, W.S. A three-dimensional model of the yeast genome. Nature 2010, 465, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Sharakhov, I.V.; Vasserlauf, I.E.; Stegnii, V.N. The features of the polytene chromosome attachment to the nuclear envelope of ovarian pseudonurse cells in Drosophila melanogaster. Genetika 1997, 33, 189–195. [Google Scholar] [PubMed]
- Hartl, T.A.; Smith, H.F.; Bosco, G. Chromosome alignment and transvection are antagonized by condensin II. Science 2008, 322, 1384–1387. [Google Scholar] [CrossRef]
- Artemov, G.; Bondarenko, S.; Sapunov, G.; Stegniy, V. Tissue-specific differences in the spatial interposition of X-chromosome and 3R chromosome regions in the malaria mosquito Anopheles messeae Fall. PLoS ONE 2015, 10, e0115281. [Google Scholar] [CrossRef] [Green Version]
- Bolzer, A.; Kreth, G.; Solovei, I.; Koehler, D.; Saracoglu, K.; Fauth, C.; Muller, S.; Eils, R.; Cremer, C.; Speicher, M.R.; et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 2005, 3, 826–842. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.L.; Murphy, C.; Buszczak, M.; Clatterbuck, S.; Goodman, R.; Gall, J.G. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006, 172, 875–884. [Google Scholar] [CrossRef]
- Tanizawa, H.; Iwasaki, O.; Tanaka, A.; Capizzi, J.R.; Wickramasinghe, P.; Lee, M.; Fu, Z.; Noma, K. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 2010, 38, 8164–8177. [Google Scholar] [CrossRef]
- Tjong, H.; Gong, K.; Chen, L.; Alber, F. Physical tethering and volume exclusion determine higher-order genome organization in budding yeast. Genome Res. 2012, 22, 1295–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avsaroglu, B.; Bronk, G.; Gordon-Messer, S.; Ham, J.; Bressan, D.A.; Haber, J.E.; Kondev, J. Effect of chromosome tethering on nuclear organization in yeast. PLoS ONE 2014, 9, e102474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osborne, C.S.; Chakalova, L.; Brown, K.E.; Carter, D.; Horton, A.; Debrand, E.; Goyenechea, B.; Mitchell, J.A.; Lopes, S.; Reik, W.; et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004, 36, 1065–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, D.A.; Nolan, T.; Fischer, B.; Pinder, A.; Crisanti, A.; Russell, S. A comprehensive gene expression atlas of sex- and tissue-specificity in the malaria vector, Anopheles gambiae. BMC Genom. 2011, 12, 296. [Google Scholar] [CrossRef] [Green Version]
- Kamali, M.; Xia, A.; Tu, Z.; Sharakhov, I.V. A new chromosomal phylogeny supports the repeated origin of vectorial capacity in malaria mosquitoes of the Anopheles gambiae complex. PLoS Pathog. 2012, 8, e1002960. [Google Scholar] [CrossRef]
- Liang, J.; Sharakhov, I.V. Premeiotic and meiotic failures lead to hybrid male sterility in the Anopheles gambiae complex. Proc. Biol. Sci. 2019, 286, 20191080. [Google Scholar] [CrossRef] [Green Version]









| Chromosome Arm | Length of the Chromosome | Total Number of Probes | Number of 10-kb Windows | Average Probe Density/kb | Average Number of Signals/Arm |
|---|---|---|---|---|---|
| X | 24.4 Mb | 2863 | 25 | 11.5 | 20 |
| 2L | 49.4 Mb | 6843 | 50 | 13.7 | 42 |
| 3R | 53.2 Mb | 7799 | 54 | 14.4 | 39 |
| 3L | 42.0 Mb | 5973 | 42 | 13.3 | 34 |
| Species and Tissue | 2L/X | 3R/X | 3L/X | Average Autosome/X |
|---|---|---|---|---|
| An. coluzzii salivary glands | 4.28 | 4.90 | 2.98 | 4.05 |
| An. coluzzii nurse cells | 3.67 | 2.81 | 2.75 | 3.08 |
| An. gambiae salivary glands | 3.44 | 3.99 | 3.90 | 3.78 |
| An. gambiae nurse cells | 3.13 | 2.68 | 2.81 | 2.87 |
| An. merus salivary glands | 5.67 | 6.02 | 4.67 | 5.45 |
| An. merus nurse cells | 6.41 | 4.74 | 3.13 | 4.76 |
| Species and Tissue | X | 2L | 3R | 3L |
|---|---|---|---|---|
| An. coluzzii salivary glands | 3.81 | 2.87 | 2.95 | 3.09 |
| An. coluzzii nurse cells | 3.47 | 2.54 | 2.28 | 2.47 |
| An. gambiae salivary glands | 3.64 | 3.47 | 3.66 | 3.78 |
| An. gambiae nurse cells | 3.37 | 3.07 | 2.69 | 3.27 |
| An. merus salivary glands | 3.59 | 2.62 | 2.90 | 2.68 |
| An. merus nurse cells | 3.27 | 2.75 | 2.73 | 2.89 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
George, P.; Kinney, N.A.; Liang, J.; Onufriev, A.V.; Sharakhov, I.V. Three-dimensional Organization of Polytene Chromosomes in Somatic and Germline Tissues of Malaria Mosquitoes. Cells 2020, 9, 339. https://doi.org/10.3390/cells9020339
George P, Kinney NA, Liang J, Onufriev AV, Sharakhov IV. Three-dimensional Organization of Polytene Chromosomes in Somatic and Germline Tissues of Malaria Mosquitoes. Cells. 2020; 9(2):339. https://doi.org/10.3390/cells9020339
Chicago/Turabian StyleGeorge, Phillip, Nicholas A. Kinney, Jiangtao Liang, Alexey V. Onufriev, and Igor V. Sharakhov. 2020. "Three-dimensional Organization of Polytene Chromosomes in Somatic and Germline Tissues of Malaria Mosquitoes" Cells 9, no. 2: 339. https://doi.org/10.3390/cells9020339
APA StyleGeorge, P., Kinney, N. A., Liang, J., Onufriev, A. V., & Sharakhov, I. V. (2020). Three-dimensional Organization of Polytene Chromosomes in Somatic and Germline Tissues of Malaria Mosquitoes. Cells, 9(2), 339. https://doi.org/10.3390/cells9020339