Differential Transcript Profiles in Cumulus-Oocyte Complexes Originating from Pre-Ovulatory Follicles of Varied Physiological Maturity in Beef Cows
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Procedures and Synchronization of Ovulation
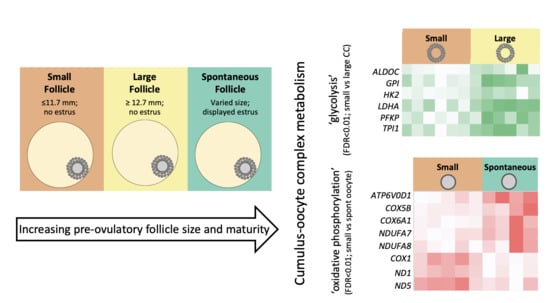
2.2. Follicle Classification
2.3. Transvaginal Aspiration for Collection of Cumulus Oocyte Complexes (COCs)
2.4. COC Recovery and Processing
2.5. Statistical Analysis of Animal Data
2.6. RNA Extraction, Library Preparation, and RNA Sequencing
2.7. RNA Sequencing Data Processing
2.8. Determination of Differentially Abundant Gene Transcripts
2.9. Gene Ontology and Pathway Enrichment Analyses
3. Results
3.1. Follicle Aspiration and Animal Data
3.2. Overview of RNA Sequencing Data
3.3. Differentially Abundant Gene Transcripts in Oocyte Pools
3.4. Differentially Abundant Gene Transcripts in Cumulus Cell Pools
4. Discussion
4.1. Cumulus Cell Transcriptome
4.2. Oocyte Transcriptome
4.3. Cumulus Cell and Oocyte Markers of Oocyte Competence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sirard, M.-A.; Richard, F.; Blondin, P.; Robert, C. Contribution of the oocyte to embryo quality. Theriogenology 2006, 65, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Fair, T.; Hulshof, S.C.J.; Hyttel, P.; Greve, T.; Boland, M. Oocyte ultrastructure in bovine primordial to early tertiary follicles. Brain Struct. Funct. 1997, 195, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Marteil, G.; Richard-Parpaillon, L.; Kubiak, J.Z. Role of oocyte quality in meiotic maturation and embryonic development. Reprod. Biol. 2009, 9, 203–224. [Google Scholar] [CrossRef]
- Ferreira, E.; Vireque, A.; Adona, P.; Meirelles, F.; Ferriani, R.; Navarro, P.A. Cytoplasmic maturation of bovine oocytes: Structural and biochemical modifications and acquisition of developmental competence. Theriogenology 2009, 71, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Hyttel, P.; Fair, T.; Callesen, H.; Greve, T. Oocyte growth, capacitation and final maturation in cattle. Theriogenology 1997, 47, 23–32. [Google Scholar] [CrossRef]
- Hendriksen, P.; Vos, P.; Steenweg, W.; Bevers, M.; Dieleman, S. Bovine follicular development and its effect on the in vitro competence of oocytes. Theriogenology 2000, 53, 11–20. [Google Scholar] [CrossRef]
- Sirard, M.; Blondin, P. Oocyte maturation and IVF in cattle. Anim. Reprod. Sci. 1996, 42, 417–426. [Google Scholar] [CrossRef]
- Cetica, P.; Pintos, L.; Dalvit, G.; Beconi, M. Activity of key enzymes involved in glucose and triglyceride catabolism during bovine oocyte maturation in vitro. Reproduction 2002, 124, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Sutton, M.; Gilchrist, R.; Thompson, J. Effects of in-vivo and in-vitro environments on the metabolism of the cumulus-oocyte complex and its influence on oocyte developmental capacity. Hum. Reprod. Updat. 2003, 9, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Sutton-McDowall, M.L.; Gilchrist, R.B.; Thompson, J.G. The pivotal role of glucose metabolism in determining oocyte developmental competence. Reproduction 2010, 139, 685–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macaulay, A.D.; Gilbert, I.; Caballero, J.; Barreto, R.; Fournier, E.; Tossou, P.; Sirard, M.-A.; Clarke, H.J.; Khandjian, É.W.; Richard, F.J.; et al. The gametic synapse: RNA transfer to the bovine oocyte. Biol. Reprod. 2014, 91, 90. [Google Scholar] [CrossRef]
- Macaulay, A.D.; Gilbert, I.; Scantland, S.; Fournier, E.; Ashkar, F.; Bastien, A.; Saadi, H.A.S.; Gagné, D.; Sirard, M.-A.; Khandjian, É.W.; et al. Cumulus cell transcripts transit to the bovine oocyte in preparation for maturation. Biol. Reprod. 2016, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigglesworth, K.; Lee, K.-B.; O’Brien, M.J.; Peng, J.; Matzuk, M.M.; Eppig, J.J. Bidirectional communication between oocytes and ovarian follicular somatic cells is required for meiotic arrest of mammalian oocytes. Proc. Natl. Acad. Sci. USA 2013, 110, E3723–E3729. [Google Scholar] [CrossRef] [Green Version]
- Arlotto, T.; Schwartz, J.-L.; First, N.; Leibfried-Rutledge, M. Aspects of follicle and oocyte stage that affect in vitro maturation and development of bovine oocytes. Theriogenology 1996, 45, 943–956. [Google Scholar] [CrossRef]
- Mamo, S.; Carter, F.; Lonergan, P.; Leal, C.L.; Al Naib, A.; Mcgettigan, P.; Mehta, J.P.; Evans, A.C.; Fair, T. Sequential analysis of global gene expression profiles in immature and in vitro matured bovine oocytes: Potential molecular markers of oocyte maturation. BMC Genom. 2011, 12, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fair, T.; Hyttel, P.; Greve, T. Bovine oocyte diameter in relation to maturational competence and transcriptional activity. Mol. Reprod. Dev. 1995, 42, 437–442. [Google Scholar] [CrossRef]
- Mourot, M.; Dufort, I.; Gravel, C.; Algriany, O.; Dieleman, S.; Sirard, M.-A. The influence of follicle size, FSH-enriched maturation medium, and early cleavage on bovine oocyte maternal mRNA levels. Mol. Reprod. Dev. 2006, 73, 1367–1379. [Google Scholar] [CrossRef]
- Atkins, J.A.; Smith, M.F.; MacNeil, M.D.; Jinks, E.M.; Abreu, F.M.; Alexander, L.J.; Geary, T.W. Pregnancy establishment and maintenance in cattle. J. Anim. Sci. 2013, 91, 722–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinks, E.M.; Smith, M.F.; Atkins, J.A.; Pohler, K.G.; Perry, G.; MacNeil, M.D.; Roberts, A.; Waterman, R.C.; Alexander, L.J.; Geary, T.W. Preovulatory estradiol and the establishment and maintenance of pregnancy in suckled beef cows. J. Anim. Sci. 2013, 91, 1176–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, G.A.; Smith, M.F.; Lucy, M.C.; Green, J.; Parks, T.E.; MacNeil, M.D.; Roberts, A.; Geary, T.W. Relationship between follicle size at insemination and pregnancy success. Proc. Natl. Acad. Sci. USA 2005, 102, 5268–5273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciernia, L.; Perry, G.; Smith, M.; Rich, J.; Northrop, E.; Perkins, S.; Green, J.; Zezeski, A.; Geary, T. Effect of estradiol preceding and progesterone subsequent to ovulation on proportion of postpartum beef cows pregnant. Anim. Reprod. Sci. 2021, 227, 106723. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, A.; Patel, O.V.; Lee, K.-B.; Park, K.-E.; Salem, M.; Yao, J.; Ireland, J.J.; Smith, G.W. Identification of novel bovine cumulus cell molecular markers predictive of oocyte competence: Functional and diagnostic implications. Biol. Reprod. 2008, 79, 301–309. [Google Scholar] [CrossRef]
- Assidi, M.; Dufort, I.; Ali, A.; Hamel, M.; Algriany, O.; Dielemann, S.; Sirard, M.-A. Identification of potential markers of oocyte competence expressed in bovine cumulus cells matured with follicle-stimulating hormone and/or phorbol myristate acetate in vitro. Biol. Reprod. 2008, 79, 209–222. [Google Scholar] [CrossRef] [Green Version]
- Kussano, N.; Leme, L.; Guimarães, A.; Franco, M.; Dode, M. Molecular markers for oocyte competence in bovine cumulus cells. Theriogenology 2016, 85, 1167–1176. [Google Scholar] [CrossRef]
- Assidi, M.; Dieleman, S.J.; Sirard, M.-A. Cumulus cell gene expression following the LH surge in bovine preovulatory follicles: Potential early markers of oocyte competence. Reproduction 2010, 140, 835–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemcova, L.; Jansova, D.; Vodickova-Kepkova, K.; Vodička, P.; Jeseta, M.; Machatkova, M.; Kanka, J. Detection of genes associated with developmental competence of bovine oocytes. Anim. Reprod. Sci. 2016, 166, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Angulo, L.; Guyader-Joly, C.; Auclair, S.; Hennequet-Antier, C.; Papillier, P.; Boussaha, M.; Fritz, S.; Hugot, K.; Moreews, F.; Ponsart, C.; et al. An integrated approach to bovine oocyte quality: From phenotype to genes. Reprod. Fertil. Dev. 2016, 28, 1276–1287. [Google Scholar] [CrossRef]
- Patel, O.V.; Bettegowda, A.; Ireland, J.J.; Coussens, P.M.; Lonergan, P.; Smith, G.W. Functional genomics studies of oocyte competence: Evidence that reduced transcript abundance for follistatin is associated with poor developmental competence of bovine oocytes. Reproduction 2007, 133, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Balboula, A.; Yamanaka, K.; Sakatani, M.; Hegab, A.; Zaabel, S.; Takahashi, M. Cathepsin B activity is related to the quality of bovine cumulus oocyte complexes and its inhibition can improve their developmental competence. Mol. Reprod. Dev. 2010, 77, 439–448. [Google Scholar] [CrossRef]
- Whitman, R.W. Weight Change, Body Condition and Beef-Cow Reproduction. Ph.D. Thesis, Colorado State University, Fort Collins, CO, USA, 1975. [Google Scholar]
- Larson, J.E.; Lamb, G.C.; Stevenson, J.S.; Johnson, S.K.; Day, M.L.; Geary, T.W.; Kesler, D.J.; DeJarnette, J.M.; Schrick, F.N.; DiCostanzo, A.; et al. Synchronization of estrus in suckled beef cows for detected estrus and artificial insemination and timed artificial insemination using gonadotropin-releasing hormone, prostaglandin F2α, and progesterone. J. Anim. Sci. 2006, 84, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G. Fqtrim: v0. 9.4 Release; Zenodo: Geneva, Switzerland, 2015. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flicek, P.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2014. Nucleic Acids Res. 2014, 42, D749–D755. [Google Scholar] [CrossRef] [PubMed]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biase, F.H.; Kimble, K.M. Functional signaling and gene regulatory networks between the oocyte and the surrounding cumulus cells. BMC Genom. 2018, 19, 1–13. [Google Scholar] [CrossRef]
- Dickinson, S.E.; Griffin, B.A.; Elmore, M.F.; Kriese-Anderson, L.; Elmore, J.B.; Dyce, P.W.; Rodning, S.P.; Biase, F.H. Transcriptome profiles in peripheral white blood cells at the time of artificial insemination discriminate beef heifers with different fertility potential. BMC Genom. 2018, 19, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sham, P.C.; Purcell, S.M. Statistical power and significance testing in large-scale genetic studies. Nat. Rev. Genet. 2014, 15, 335–346. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moorey, S.E.; Walker, B.N.; Elmore, M.F.; Elmore, J.B.; Rodning, S.P.; Biase, F.H. Rewiring of gene expression in circulating white blood cells is associated with pregnancy outcome in heifers (Bos taurus). Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- The Gene Ontology Consortium; Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology resource: Enriching a gold mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navale, A.M.; Paranjape, A.N. Glucose transporters: Physiological and pathological roles. Biophys. Rev. 2016, 8, 5–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimoto, H.; Matsutani, R.; Yamamoto, S.; Takahashi, T.; Hayashi, K.-G.; Miyamoto, A.; Hamano, S.; Tetsuka, M. Gene expression of glucose transporter (GLUT) 1, 3 and 4 in bovine follicle and corpus luteum. J. Endocrinol. 2006, 188, 111–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boruszewska, D.; Sinderewicz, E.; Kowalczyk-Zieba, I.; Grycmacher, K.; Woclawek-Potocka, I. The effect of lysophosphatidic acid during in vitro maturation of bovine Cumulus-Oocyte complexes: Cumulus expansion, glucose metabolism and expression of genes involved in the ovulatory cascade, oocyte and blastocyst competence. Reprod. Biol. Endocrinol. 2015, 13, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Chi, M.M.; Schedl, T.; Moley, K.H. An intercellular pathway for glucose transport into mouse oocytes. Am. J. Physiol. Metab. 2012, 302, E1511–E1518. [Google Scholar] [CrossRef] [Green Version]
- Krisher, R.L.; Bavister, B.D. Enhanced glycolysis after maturation of bovine oocytes in vitro is associated with increased developmental competence. Mol. Reprod. Dev. 1999, 53, 19–26. [Google Scholar] [CrossRef]
- Thompson, J.G.; Gilchrist, R.; Sutton-McDowall, M.L. The metabolism of the ruminant cumulus-oocyte complex revisited. Biosci. Proc. 2019, 2014, 311–326. [Google Scholar] [CrossRef]
- Saito, T.; Hiroi, M.; Kato, T. Development of Glucose Utilization Studied in Single Oocytes and Preimplantation Embryos from Mice. Biol. Reprod. 1994, 50, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.E.; Adriaens, I.; Leese, H.J.; Gosden, R.G.; Picton, H.M. Carbohydrate metabolism by murine ovarian follicles and oocytes grown in vitro. Reproduction 2007, 134, 415–424. [Google Scholar] [CrossRef] [Green Version]
- Krisher, R.L. The effect of oocyte quality on development. J. Anim. Sci. 2004, 82 (Suppl. 13), E14–E23. [Google Scholar]
- Shen, Q.; Chen, M.; Zhao, X.; Liu, Y.; Ren, X.; Zhang, L. Versican expression level in cumulus cells is associated with human oocyte developmental competence. Syst. Biol. Reprod. Med. 2020, 66, 176–184. [Google Scholar] [CrossRef]
- Cetica, P.D.; Pintos, L.N.; Dalvit, G.C.; Beconi, M.T. Effect of lactate dehydrogenase activity and isoenzyme localization in bovine oocytes and utilization of oxidative substrates on in vitro maturation. Theriogenology 1999, 51, 541–550. [Google Scholar] [CrossRef]
- Sugiura, K.; Pendola, F.L.; Eppig, J.J. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: Energy metabolism. Dev. Biol. 2005, 279, 20–30. [Google Scholar] [CrossRef] [Green Version]
- Sugiura, K.; Eppig, J.J. Society for Reproductive Biology Founders’ Lecture 2005. Control of metabolic cooperativity between oocytes and their companion granulosa cells by mouse oocytes. Reprod. Fertil. Dev. 2005, 17, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, K.; Su, Y.-Q.; Diaz, F.J.; Pangas, S.A.; Sharma, S.; Wigglesworth, K.; O’Brien, M.J.; Matzuk, M.M.; Shimasaki, S.; Eppig, J.J. Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development 2007, 134, 2593–2603. [Google Scholar] [CrossRef] [Green Version]
- Ornitz, D.M.; Itoh, N. The fibroblast growth factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krisher, R.L.; Prather, R.S. A role for the Warburg effect in preimplantation embryo development: Metabolic modification to support rapid cell proliferation. Mol. Reprod. Dev. 2012, 79, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Bertoldo, M.; Listijono, D.R.; Ho, W.-H.J.; Riepsamen, A.H.; Goss, D.M.; Richani, D.; Jin, X.L.; Mahbub, S.; Campbell, J.M.; Habibalahi, A.; et al. NAD+ repletion rescues female fertility during reproductive aging. Cell Rep. 2020, 30, 1670–1681.e7. [Google Scholar] [CrossRef] [Green Version]
- Ullah, M.S.; Davies, A.J.; Halestrap, A.P. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1α-dependent mechanism. J. Biol. Chem. 2006, 281, 9030–9037. [Google Scholar] [CrossRef] [Green Version]
- Chappel, S. The role of mitochondria from mature oocyte to viable blastocyst. Obstet. Gynecol. Int. 2013, 2013, 183024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Blerkom, J.; Davis, P.W.; Lee, J. Fertilization and early embryology: ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum. Reprod. 1995, 10, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Stojkovic, M.; Machado, S.A.; Stojkovic, P.; Zakhartchenko, V.; Hutzler, P.; Gonçalves, P.B.; Wolf, E. Mitochondrial distribution and adenosine triphosphate content of bovine oocytes before and after in vitro maturation: Correlation with morphological criteria and developmental capacity after in vitro fertilization and culture. Biol. Reprod. 2001, 64, 904–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martazanova, B.; Mishieva, N.; Bogatyreva, K.; Veyukova, M.; Korolkova, A.; Burmenskaya, O.; Abubakirov, A. Versican (VCAN) mRNA expression in cumulus cells predicts embryo quality, but does not predict pregnancy outcome in patients with reduced ovarian reserve. Fertil. Steril. 2018, 110, e310. [Google Scholar] [CrossRef]
- Larsen, W.J.; Wert, S.E.; Brunner, G.D. A dramatic loss of cumulus cell gap junctions is correlated with germinal vesicle breakdown in rat oocytes. Dev. Biol. 1986, 113, 517–521. [Google Scholar] [CrossRef]
- Vanderhyden, B.C.; Armstrong, D.T. Role of cumulus cells and serum on the in vitro maturation, fertilization, and subsequent development of rat oocytes. Biol. Reprod. 1989, 40, 720–728. [Google Scholar] [CrossRef] [Green Version]
- Gutnisky, C.; Dalvit, G.C.; Pintos, L.N.; Thompson, J.G.; Beconi, M.T.; Cetica, P.D. Influence of hyaluronic acid synthesis and cumulus mucification on bovine oocyte in vitro maturation, fertilisation and embryo development. Reprod. Fertil. Dev. 2007, 19, 488–497. [Google Scholar] [CrossRef]
- Dorey, K.; Amaya, E. FGF signalling: Diverse roles during early vertebrate embryogenesis. Development 2010, 137, 3731–3742. [Google Scholar] [CrossRef] [Green Version]
- Fields, S.D.; Hansen, P.J.; Ealy, A.D. Fibroblast growth factor requirements for in vitro development of bovine embryos. Theriogenology 2011, 75, 1466–1475. [Google Scholar] [CrossRef]
- Du, C.; Davis, J.S.; Chen, C.; Li, Z.; Cao, Y.; Sun, H.; Shao, B.-S.; Lin, Y.-X.; Wang, Y.-S.; Yang, L.-G.; et al. FGF2/FGFR signaling promotes Cumulus-Oocyte complex maturation in vitro. Reproduction 2021, 161, 205–214. [Google Scholar] [CrossRef]
- Melo, E.O.; Cordeiro, D.M.; Pellegrino, R.; Wei, Z.; Daye, Z.J.; Nishimura, R.C.; Dode, M.A.N. Identification of molecular markers for oocyte competence in bovine cumulus cells. Anim. Genet. 2016, 48, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Iwata, H.; Goto, H.; Tanaka, H.; Sakaguchi, Y.; Kimura, K.; Kuwayama, T.; Monji, Y. Effect of maternal age on mitochondrial DNA copy number, ATP content and IVF outcome of bovine oocytes. Reprod. Fertil. Dev. 2011, 23, 424–432. [Google Scholar] [CrossRef]
- John, J.S. The control of mtDNA replication during differentiation and development. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 1345–1354. [Google Scholar] [CrossRef]
- Biggers, J.D.; Whittingham, D.G.; Donahue, R.P. The pattern of energy metabolism in the mouse oocyte and zygote. Proc. Nat. Acad. Sci. USA 1967, 58, 560–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.T.; Freeman, E.A.; Gardner, D.; Hunt, P.A. Oxidative metabolism of pyruvate is required for meiotic maturation of murine oocytes in vivo. Biol. Reprod. 2007, 77, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.S.; Wohlgemuth, S.; Tribulo, P.; Siqueira, L.G.B.; Null, D.J.; Cole, J.B.; Da Silva, M.V.; Hansen, P.J. A single nucleotide polymorphism in COQ9 affects mitochondrial and ovarian function and fertility in Holstein cows. Biol. Reprod. 2017, 96, 652–663. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.M.; Chiaratti, M.R.; Macabelli, C.H.; Rodrigues, C.A.; Ferraz, M.L.; Watanabe, Y.F.; Smith, L.C.; Meirelles, F.V.; Baruselli, P.S. The infertility of repeat-breeder cows during summer is associated with decreased mitochondrial DNA and increased expression of mitochondrial and apoptotic genes in oocytes. Biol. Reprod. 2016, 94, 66. [Google Scholar] [CrossRef]
- May-Panloup, P.; Chrétien, M.; Jacques, C.; Vasseur, C.; Malthièry, Y.; Reynier, P. Low oocyte mitochondrial DNA content in ovarian insufficiency. Hum. Reprod. 2005, 20, 593–597. [Google Scholar] [CrossRef] [Green Version]
- Reynier, P.; May-Panloup, P.; Chretien, M.-F.; Morgan, C.; Jean, M.; Savagner, F.; Barriere, P.; Malthiery, Y. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol. Hum. Reprod. 2001, 7, 425–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Shourbagy, S.H.; Spikings, E.C.; Freitas, M.; John, J.C.S. Mitochondria directly influence fertilisation outcome in the pig. Reproduction 2006, 131, 233–245. [Google Scholar] [CrossRef] [Green Version]
- D’Souza, A.R.; Minczuk, M. Mitochondrial transcription and translation: Overview. Essays Biochem. 2018, 62, 309–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexeyev, M.F. Mitochondrial transcription in mammalian cells. Front. Biosci. 2017, 22, 835–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondadori, R.; Neves, J.; Gonçalves, P. Protein kinase C (PKC) role in bovine oocyte maturation and early embryo development. Anim. Reprod. Sci. 2008, 107, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Fana, H.-Y.; Tonga, C.; Lia, M.-Y.; Liana, L.; Chena, D.-Y.; Schattenb, H.; Sun, Q.-Y. Translocation of the classic protein kinase C isoforms in porcine oocytes: Implications of protein kinase C involvement in the regulation of nuclear activity and cortical granule exocytosis. Exp. Cell Res. 2002, 277, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-Q.; Zhang, X.; Li, X.-H.; Cheng, H.-H.; Kuai, Y.-R.; Wang, S.; Guo, Y.-L. Translocation of classical PKC and cortical granule exocytosis of human oocyte in germinal vesicle and metaphase II stage. Acta Pharmacol. Sin. 2006, 27, 1353–1358. [Google Scholar] [CrossRef]
- Feitosa, W.B.; Lopes, E.; Visintin, J.A.; Assumpção, M.E.O.D. Endoplasmic reticulum distribution during bovine oocyte activation is regulated by protein kinase C via actin filaments. J. Cell. Physiol. 2020, 235, 5823–5834. [Google Scholar] [CrossRef]
- Dumollard, R.; Duchen, M.; Sardet, C. Calcium signals and mitochondria at fertilisation. Semin. Cell Dev. Biol. 2006, 17, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Pohler, K.G. Effect of Ovulatory Follicle Size on Steroidogenic Capacity, Molecular Markers of Oocyte Competence and Bovine Pregnancy Associated with Glycoproteins in Beef Cows. Ph.D. Thesis, University of Missouri, Columbia, MO, USA, 2011. [Google Scholar]



| Parameter | Small Follicle Classification | Large Follicle Classification | Spontaneous Follicle Classification |
|---|---|---|---|
| Follicle size at GnRH2 a | 10.4 x ± 0.1 mm (8.5–11.7 mm) | 13.6 y ± 0.1 mm (12.7–15.3 mm) | 12.2 z ± 0.2 mm (11.7–14.0 mm) |
| Cow age b | 6.5 ± 0.4 years (4–12 years) | 6.3 ± 0.4 years (4–9 years) | 6.9 ± 0.5 years (4–13 years) |
| Cow weight c | 548 ± 11 kg (454–674 kg) | 564 ± 11 kg (452–668 kg) | 548 ± 14 kg (468–646 kg) |
| Cow BCS d | 4.8 ± 0.1 (4–5) | 4.8 ± 0.1 (4–6) | 4.7 ± 0.1 (4–5) |
| Cow days postpartum e | 88 ± 1.7 days (65–96 days) | 86 ± 1.7 days (58–98 days) | 86 ± 2.0 days (76–95 days) |
| Time from PGF to GnRH2 f | 51 ± 1.5 h (43–56 h) | 51 ± 1.5 h (43–56 h) | NA |
| Time from GnRH2 or estrous onset to follicle aspiration g | 23 ± 0.8 h (18–31 h) | 22 ± 0.6 h (17–30 h) | 21 ± 0.7 h (17–24 h) |
| Time from follicle aspiration to snap freezing of samples h | 22 ± 1.6 min (11–45 min) | 24 ± 1.6 min (11–34 min) | 25 ± 2.0 min (12–45 min) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moorey, S.E.; Monnig, J.M.; Smith, M.F.; Ortega, M.S.; Green, J.A.; Pohler, K.G.; Bridges, G.A.; Behura, S.K.; Geary, T.W. Differential Transcript Profiles in Cumulus-Oocyte Complexes Originating from Pre-Ovulatory Follicles of Varied Physiological Maturity in Beef Cows. Genes 2021, 12, 893. https://doi.org/10.3390/genes12060893
Moorey SE, Monnig JM, Smith MF, Ortega MS, Green JA, Pohler KG, Bridges GA, Behura SK, Geary TW. Differential Transcript Profiles in Cumulus-Oocyte Complexes Originating from Pre-Ovulatory Follicles of Varied Physiological Maturity in Beef Cows. Genes. 2021; 12(6):893. https://doi.org/10.3390/genes12060893
Chicago/Turabian StyleMoorey, Sarah E., Jenna M. Monnig, Michael F. Smith, M. Sofia Ortega, Jonathan A. Green, Ky G. Pohler, G. Alan Bridges, Susanta K. Behura, and Thomas W. Geary. 2021. "Differential Transcript Profiles in Cumulus-Oocyte Complexes Originating from Pre-Ovulatory Follicles of Varied Physiological Maturity in Beef Cows" Genes 12, no. 6: 893. https://doi.org/10.3390/genes12060893
APA StyleMoorey, S. E., Monnig, J. M., Smith, M. F., Ortega, M. S., Green, J. A., Pohler, K. G., Bridges, G. A., Behura, S. K., & Geary, T. W. (2021). Differential Transcript Profiles in Cumulus-Oocyte Complexes Originating from Pre-Ovulatory Follicles of Varied Physiological Maturity in Beef Cows. Genes, 12(6), 893. https://doi.org/10.3390/genes12060893