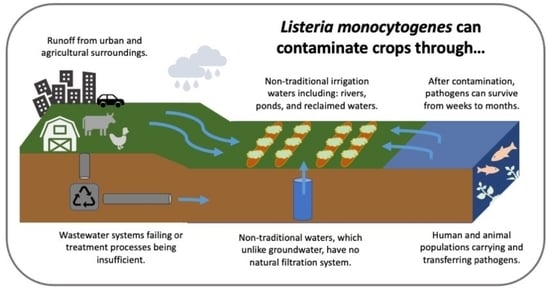
Listeria monocytogenes in Irrigation Water: An Assessment of Outbreaks, Sources, Prevalence, and Persistence
Abstract
:1. Introduction
2. L. monocytogenes—Foodborne Pathogen and Produce Outbreaks
3. Presence and Prevalence of L. monocytogenes in Water and Natural Environments
4. Listeria monocytogenes and Overall Pathogen Presence: Reclaimed and Recycled Waters
5. Persistence of Listeria spp. in Water, Soil, and on Crop Surfaces
6. Implication of Internalin A in L. monocytogenes
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- U.S. Geological Survey Water Questions & Answers. Available online: https://water.usgs.gov/edu/qa-usage-gw.html (accessed on 2 September 2019).
- U.S. Geological Survey Ground Water Decline and Depletion. Available online: https://www.usgs.gov/special-topic/water-science-school/science/groundwater-decline-and-depletion?qt-science_center_objects=0#qt-science_center_objects (accessed on 1 September 2019).
- U.S. Department of Agriculture Water for Food Production Systems Challenge Area Fiscal Year 2017. Available online: https://nifa.usda.gov/sites/default/files/rfa/FY2017_AFRI_Water_for%20Food%20Production%20Systems.pdf (accessed on 19 August 2018).
- Dery, J.L.; Suri, M.R.; Brassill, N.; Pee, D.; Goeringer, P.; Sapkota, A.R.; Rock, C.; Rosenberg-Goldstein, R.E. Recycled Water and Related Terms Relevant for Agriculture. Available online: http://conservewaterforfood.org/extension/ (accessed on 15 August 2019).
- U.S. National Ground Water Association Facts about Global Groundwater Usage. Available online: https://www.ngwa.org/what-is-groundwater/About-groundwater/facts-about-global-groundwater-usage (accessed on 2 September 2019).
- Fisher, N.; Bourne, A.; Plunkett, D. Outbreak Alert! 2015: A Review of Foodborne Illnesses in the US from 2004–2013; Center for Science in the Public Interest: Washington, DC, USA, 2015. [Google Scholar]
- Painter, J.A.; Hoekstra, R.M.; Ayers, T.; Tauxe, R.V.; Braden, C.R.; Angulo, F.J.; Griffin, P.M. Attribution of Foodborne Illnesses, Hospitalizations, and Deaths to Food Commodities by Using Outbreak Data, United States, 1998–2008. Emerg. Infect. Dis. 2013, 19, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Ijabadeniyi, O.A.; Debusho, L.K.; Vanderlinde, M.; Buys, E.M. Irrigation water as a potential preharvest source of bacterial contamination of vegetables. J. Food Saf. 2011, 31, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Steele, M.; Odumeru, J. Irrigation Water as Source of Foodborne Pathogens on Fruit and Vegetables. J. Food Prot. 2004, 67, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Cooley, M.B.; Quiñones, B.; Oryang, D.; Mandrell, R.E.; Gorski, L. Prevalence of Shiga Toxin Producing Escherichia Coli, Salmonella Enterica, and Listeria Monocytogenes at Public Access Watershed Sites in a California Central Coast Agricultural Region. Front. Cell. Infect. Microbiol. 2014, 4, 30. [Google Scholar] [CrossRef] [Green Version]
- Strawn, L.K.; Gröhn, Y.T.; Warchocki, S.; Worobo, R.W.; Bihn, E.A.; Wiedmann, M. Risk Factors Associated with Salmonella and Listeria Monocytogenes Contamination of Produce Fields. Appl. Environ. Microbiol. 2013, 79, 7618–7627. [Google Scholar] [CrossRef] [Green Version]
- Strawn, L.K.; Fortes, E.D.; Bihn, E.A.; Nightingale, K.K.; Gröhn, Y.T.; Worobo, R.W.; Wiedmann, M.; Bergholz, P.W. Landscape and Meteorological Factors Affecting Prevalence of Three Food-Borne Pathogens in Fruit and Vegetable Farms. Appl. Environ. Microbiol. 2013, 79, 588–600. [Google Scholar] [CrossRef] [Green Version]
- Pachepsky, Y.; Morrow, J.; Guber, A.; Shelton, D.; Rowland, R.; Davies, G. Effect of Biofilm in Irrigation Pipes on Microbial Quality of Irrigation Water. Lett. Appl. Microbiol. 2012, 54, 217–224. [Google Scholar] [CrossRef]
- Materon, L.A.; Martinez-Garcia, M.; McDonald, V. Identification of Sources of Microbial Pathogens on Cantaloupe Rinds from Pre-Harvest to Post-Harvest Operations. World J. Microbiol. Biotechnol. 2007, 23, 1281–1287. [Google Scholar] [CrossRef]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasieqicz, M.J. Listeria Monocytogenes Persistence in Food-Associated Environments: Epidemiology, Strain Characteristics, and Implications for Public Health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Gião, M.S.; Keevil, C.W. Listeria Monocytogenes Can Form Biofilms in Tap Water and Enter Into the Viable but Non-Cultivable State. Microb. Ecol. 2014, 67, 603–611. [Google Scholar] [CrossRef]
- Mañas, P.; Castro, E.; de las Heras, J. Irrigation with Treated Wastewater: Effects on Soil, Lettuce (Lactuca Sativa L.) Crop and Dynamics of Microorganisms. J. Environ. Sci. Health Part A 2009, 44, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Manuel, C.S.; van Stelten, A.; Wiedmann, M.; Nightingale, K.K.; Orsi, R.H. Prevalence and Distribution of Listeria Monocytogenes InlA Alleles Prone to Phase Variation and InlA Alleles with Premature Stop Codon Mutations among Human, Food, Animal, and Environmental Isolates. Appl. Environ. Microbiol. 2015, 81, 8339–8345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorski, L.; Parker, C.T.; Liang, A.S.; Walker, S.; Romanolo, K.F. The Majority of Genotypes of the Virulence Gene InlA Are Intact among Natural Watershed Isolates of Listeria Monocytogenes from the Central California Coast. PLoS ONE 2016, 11, e0167566. [Google Scholar] [CrossRef] [Green Version]
- Weller, D.; Wiedmann, M.; Strawn, L.K. Irrigation Is Significantly Associated with an Increased Prevalence of Listeria Monocytogenes in Produce Production Environments in New York State. J. Food Prot. 2015, 78, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Szonyi, B.; Gautam, R.; Nightingale, K.; Ancis, J.; Ivanek, R. Risk Factors for Microbial Contamination in Fruits and Vegetables at the Preharvest Level: A Systematic Review. J. Food Prot. 2012, 75, 2055–2081. [Google Scholar] [CrossRef] [PubMed]
- Guzel, M.; Moreira, R.G.; Omac, B.; Castell-Perez, M.E. Quantifying the Effectiveness of Washing Treatments on the Microbial Quality of Fresh-Cut Romaine Lettuce and Cantaloupe. LWT 2017, 86, 270–276. [Google Scholar] [CrossRef]
- MacGowan, A.P.; Bowker, K.; McLauchlin, J.; Bennett, P.M.; Reeves, D.S. The Occurrence and Seasonal Changes in the Isolation of Listeria Spp. in Shop Bought Food Stuffs, Human Faeces, Sewage and Soil from Urban Sources. Int. J. Food Microbiol. 1994, 21, 325–334. [Google Scholar] [CrossRef]
- Stea, E.C.; Purdue, L.M.; Jamieson, R.C.; Yost, C.K.; Truelstrup Hansen, L. Comparison of the Prevalences and Diversities of Listeria Species and Listeria Monocytogenes in an Urban and a Rural Agricultural Watershed. Appl. Environ. Microbiol. 2015, 81, 3812–3822. [Google Scholar] [CrossRef] [Green Version]
- Liao, J.; Guo, X.; Weller, D.L.; Pollak, S.; Buckley, D.H.; Wiedmann, M.; Cordero, O.X. Nationwide Genomic Atlas of Soil-Dwelling Listeria Reveals Effects of Selection and Population Ecology on Pangenome Evolution. Nat. Microbiol. 2021, 6, 1021–1030. [Google Scholar] [CrossRef]
- Cossart, P. Invasion of Mammalian Cells by Listeria Monocytogenes: Functional Mimicry to Subvert Cellular Functions. Trends Cell Biol. 2003, 13, 23–31. [Google Scholar] [CrossRef]
- U.S. Centers for Disease Control and Prevention Listeria (Listeriosis). Available online: https://www.cdc.gov/listeria/index.html (accessed on 16 April 2022).
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.R.; Salter, M.; Tarr, C.; Conrad, A.; Harvey, E.; Steinbock, L.; Saupe, A.; Sorenson, A.; Katz, L.; Stroika, S.; et al. Notes from the Field: Listeriosis Associated with Stone Fruit—United States, 2014. Morb. Mortal. Wkly. Rep. 2015, 64, 282–283. [Google Scholar]
- Self, J.L.; Conrad, A.; Stroika, S.; Jackson, A.; Whitlock, L.; Jackson, K.A.; Beal, J.; Wellman, A.; Fatica, M.K.; Bidol, S.; et al. Multistate Outbreak of Listeriosis Associated with Packaged Leafy Green Salads, United States and Canada, 2015–2016. Emerg. Infect. Dis. 2019, 25, 1461–1468. [Google Scholar] [CrossRef]
- Botzler, R.G.; Cowan, A.B.; Wetzler, T.F. SURVIVAL OF Listeria Monocytogenes IN SOIL AND WATER. J. Wildl. Dis. 1974, 10, 204–212. [Google Scholar] [CrossRef]
- Chapin, T.K.; Nightingale, K.K.; Worobo, R.W.; Wiedmann, M.; Strawn, L.K. Geographical and Meteorological Factors Associated with Isolation of Listeria Species in New York State Produce Production and Natural Environments. J. Food Prot. 2014, 77, 1919–1928. [Google Scholar] [CrossRef]
- Colburn, K.G.; Kaysner, C.A.; Abeyta, C.; Wekell, M.M. Listeria Species in a California Coast Estuarine Environment. Appl. Environ. Microbiol. 1990, 56, 2007–2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowe, M.J.; Jackson, E.D.; Mori, J.G.; Bell, C.R. Listeria Monocytogenes Survival in Soil and Incidence in Agricultural Soils†. J. Food Prot. 1997, 60, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Fenlon, D.R. Listeria Monocytogenes in the Natural Environment. In Listeria, Listeriosis, and Food Safety; Ryser, E.T., Marth, E.H., Eds.; Marcel Dekker: New York, NY, USA, 1999; pp. 21–38. [Google Scholar]
- Ivanek, R.; Gröhn, Y.T.; Wells, M.T.; Lembo, A.J.; Sauders, B.D.; Wiedmann, M. Modeling of Spatially Referenced Environmental and Meteorological Factors Influencing the Probability of Listeria Species Isolation from Natural Environments. Appl. Environ. Microbiol. 2009, 75, 5893–5909. [Google Scholar] [CrossRef] [Green Version]
- Lang-Halter, E.; Schober, S.; Scherer, S. Permanent Colonization of Creek Sediments, Creek Water and Limnic Water Plants by Four Listeria Species in Low Population Densities. Z. Nat. C 2016, 71, 335–345. [Google Scholar] [CrossRef]
- Linke, K.; Rückerl, I.; Brugger, K.; Karpiskova, R.; Walland, J.; Muri-Klinger, S.; Tichy, A.; Wagner, M.; Stessl, B. Reservoirs of Listeria Species in Three Environmental Ecosystems. Appl. Environ. Microbiol. 2014, 80, 5583–5592. [Google Scholar] [CrossRef] [Green Version]
- Locatelli, A.; Depret, G.; Jolivet, C.; Henry, S.; Dequiedt, S.; Piveteau, P.; Hartmann, A. Nation-Wide Study of the Occurrence of Listeria Monocytogenes in French Soils Using Culture-Based and Molecular Detection Methods. J. Microbiol. Methods 2013, 93, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, A.; Spor, A.; Jolivet, C.; Piveteau, P.; Hartmann, A. Biotic and Abiotic Soil Properties Influence Survival of Listeria Monocytogenes in Soil. PLoS ONE 2013, 8, e75969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyautey, E.; Hartmann, A.; Pagotto, F.; Tyler, K.; Lapen, D.R.; Wilkes, G.; Piveteau, P.; Rieu, A.; Robertson, W.J.; Medeiros, D.T.; et al. Characteristics and Frequency of Detection of Fecal Listeria Monocytogenes Shed by Livestock, Wildlife, and Humans. Can. J. Microbiol. 2007, 53, 1158–1167. [Google Scholar] [CrossRef]
- Lyautey, E.; Lapen, D.R.; Wilkes, G.; McCleary, K.; Pagotto, F.; Tyler, K.; Hartmann, A.; Piveteau, P.; Rieu, A.; Robertson, W.J.; et al. Distribution and Characteristics of Listeria Monocytogenes Isolates from Surface Waters of the South Nation River Watershed, Ontario, Canada. Appl. Environ. Microbiol. 2007, 73, 5401–5410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nightingale, K.K.; Windham, K.; Wiedmann, M. Evolution and Molecular Phylogeny of Listeria Monocytogenes Isolated from Human and Animal Listeriosis Cases and Foods. J. Bacteriol. 2005, 187, 5537–5551. [Google Scholar] [CrossRef] [Green Version]
- Nightingale, K.K.; Schukken, Y.H.; Nightingale, C.R.; Fortes, E.D.; Ho, A.J.; Her, Z.; Grohn, Y.T.; McDonough, P.L.; Wiedmann, M. Ecology and Transmission of Listeria Monocytogenes Infecting Ruminants and in the Farm Environment. Appl. Environ. Microbiol. 2004, 70, 4458–4467. [Google Scholar] [CrossRef] [Green Version]
- Sauders, B.D.; Overdevest, J.; Fortes, E.; Windham, K.; Schukken, Y.; Lembo, A.; Wiedmann, M. Diversity of Listeria Species in Urban and Natural Environments. Appl. Environ. Microbiol. 2012, 78, 4420–4433. [Google Scholar] [CrossRef] [Green Version]
- Sauders, B.D.; Durak, M.Z.; Fortes, E.; Windham, K.; Schukken, Y.; Lembo, A.J.; Akey, B.; Nightingale, K.K.; Wiedmann, M. Molecular Characterization of Listeria Monocytogenes from Natural and Urban Environments. J. Food Prot. 2006, 69, 93–105. [Google Scholar] [CrossRef]
- Soni, D.K.; Singh, R.K.; Singh, D.V.; Dubey, S.K. Characterization of Listeria Monocytogenes Isolated from Ganges Water, Human Clinical and Milk Samples at Varanasi, India. Infect. Genet. Evol. 2013, 14, 83–91. [Google Scholar] [CrossRef]
- Vivant, A.-L.; Garmyn, D.; Piveteau, P. Listeria Monocytogenes, a down-to-Earth Pathogen. Front. Cell. Infect. Microbiol. 2013, 3, 87. [Google Scholar] [CrossRef] [Green Version]
- Weis, J.; Seeliger, H.P.R. Incidence of Listeria Monocytogenes in Nature. Appl. Microbiol. 1975, 30, 29–32. [Google Scholar] [CrossRef]
- Wilkes, G.; Edge, T.A.; Gannon, V.P.J.; Jokinen, C.; Lyautey, E.; Neumann, N.F.; Ruecker, N.; Scott, A.; Sunohara, M.; Topp, E.; et al. Associations among Pathogenic Bacteria, Parasites, and Environmental and Land Use Factors in Multiple Mixed-Use Watersheds. Water Res. 2011, 45, 5807–5825. [Google Scholar] [CrossRef] [PubMed]
- Girardin, H.; Morris, C.E.; Albagnac, C.; Dreux, N.; Glaux, C.; Nguyen-The, C. Behaviour of the Pathogen Surrogates Listeria Innocua and Clostridium Sporogenes during Production of Parsley in Fields Fertilized with Contaminated Amendments. FEMS Microbiol. Ecol. 2005, 54, 287–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribot, E.M.; Hise, K.B. Future Challenges for Tracking Foodborne Diseases. EMBO Rep. 2016, 17, 1499–1505. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.Y.; Henao, O.L.; Griffin, P.M.; Vudia, D.J.; Cronquist, A.B.; Hurd, S.; Tobin-D’Angelo, M.; Ryan, P.; Smith, K.; Lathrop, S.; et al. Infection with Pathogens Transmitted Commonly Through Food and the Effect of Increasing Use of Culture-Independent Diagnostic Tests on Surveillance–Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2012–2015. Morb. Mortal. Wkly. Rep. 2016, 65, 368–371. [Google Scholar] [CrossRef]
- PEW Charitable Trusts as Tests for Foodborne Illness Evolve, Disease Tracking Systems Must Adapt. Available online: https://www.pewtrusts.org/en/research-and-analysis/articles/2018/04/as-tests-for-foodborne-illness-evolve-disease-tracking-systems-must-adapt (accessed on 2 September 2019).
- McCollum, J.T.; Cronquist, A.B.; Silk, B.J.; Jackson, K.A.; O’Connor, K.A.; Cosgrove, S.; Gossack, J.P.; Parachini, S.S.; Jain, N.S.; Ettestad, P.; et al. Multistate Outbreak of Listeriosis Associated with Cantaloupe. N. Engl. J. Med. 2013, 369, 944–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laksanalamai, P.; Joseph, L.A.; Silk, B.J.; Burall, L.S.; Tarr, C.L.; Gerner-Smidt, P.; Datta, A.R. Genomic Characterization of Listeria Monocytogenes Strains Involved in a Multistate Listeriosis Outbreak Associated with Cantaloupe in US. PLoS ONE 2012, 7, e42448. [Google Scholar] [CrossRef]
- U.S. Centers for Disease Control and Prevention. Multistate Outbreak of Listeriosis Associated with Jensen Farms Cantaloupe—United States, August–September 2011. MMWR Morb. Mortal. Wkly. Rep. 2011, 60, 1357–1358. [Google Scholar]
- U.S. Food and Drug Administration Wholesome Soy Products, Inc. Available online: https://www.fda.gov/about-fda/ora-foia-electronic-reading-room/wholesome-soy-products-inc (accessed on 31 July 2019).
- Angelo, K.M.; Conrad, A.R.; Saupe, A.; Dragoo, H.; West, N.; Sorenson, A.; Barnes, A.; Doyle, M.; Beal, J.; Jackson, K.A.; et al. Multistate Outbreak of Listeria Monocytogenes Infections Linked to Whole Apples Used in Commercially Produced, Prepackaged Caramel Apples: United States, 2014–2015. Epidemiol. Infect. 2017, 145, 848–856. [Google Scholar] [CrossRef] [Green Version]
- U.S. Food and Drug Administration FDA. Investigated Listeria Outbreak Linked to Frozen Vegetables. Available online: https://www.fda.gov/food/outbreaks-foodborne-illness/fda-investigated-listeria-outbreak-linked-frozen-vegetables (accessed on 1 September 2019).
- U.S. Centers for Disease Control and Prevention Listeria (Listeriosis). Available online: https://www.cdc.gov/listeria/outbreaks/enoki-mushrooms-03-20/index.html (accessed on 19 March 2022).
- Washington State Department of Health Food Recalls and Safety Alerts in Washington. Available online: https://doh.wa.gov/you-and-your-family/food-safety/recalls (accessed on 19 March 2022).
- U.S. Centers for Disease Control and Prevention Listeria (Listeriosis). Available online: https://www.cdc.gov/listeria/outbreaks/packaged-salad-12-21-b/index.html (accessed on 16 April 2022).
- U.S. Centers for Disease Control and Prevention Listeria (Listeriosis). Available online: https://www.cdc.gov/listeria/outbreaks/packaged-salad-mix-12-21/index.html (accessed on 19 March 2022).
- Beuchat, L.R.; Ryu, J.H. Produce Handling and Processing Practices. Emerg. Infect. Dis. 1997, 3, 459–465. [Google Scholar] [CrossRef] [Green Version]
- U.S. Department of Healh & Human Services FoodSafety.Gov. Available online: https://www.foodsafety.gov/recalls-and-outbreaks (accessed on 2 September 2019).
- U.S. Centers for Disease Control and Prevention Foodborne Outbreaks: Step 5: Solve Point of Contamination and Source of the Food. Available online: https://www.cdc.gov/foodsafety/outbreaks/investigating-outbreaks/investigations/index.html (accessed on 2 September 2020).
- Jenny Schell Food Poison Journal. Available online: https://www.foodpoisonjournal.com/food-poisoning-information/salads-recalled-over-listeria-fears/ (accessed on 19 June 2022).
- U.S. Food and Drug Administration. FDA Investigated Multistate Outbreak of E. Coli O157:H7 Infections Linked to Romaine Lettuce from Yuma Growing Region. Available online: https://www.fda.gov/food/outbreaks-foodborne-illness/fda-investigated-multistate-outbreak-e-coli-o157h7-infections-linked-romaine-lettuce-yuma-growing (accessed on 31 August 2019).
- U.S. Food and Drug Administration. Environmental Assessment of Factors Potentially Contributing to the Contamination of Romaine Lettuce Implicated in a Multi-State Outbreak of E. Coli O157:H7. Available online: https://www.fda.gov/food/outbreaks-foodborne-illness/environmental-assessment-factors-potentially-contributing-contamination-romaine-lettuce-implicated (accessed on 31 August 2019).
- U.S. Food and Drug Administration. Outbreak Investigation of E. Coli: Romaine (November 2018). Available online: https://www.fda.gov/food/outbreaks-foodborne-illness/outbreak-investigation-e-coli-o157h7-linked-romaine-lettuce-grown-ca?utm_campaign=Outbreak_Romaine_11262018&utm_medium=email&utm_source=Eloqua (accessed on 31 August 2019).
- Ponniah, J.; Robin, T.; Paie, M.S.; Radu, S.; Ghazali, F.M.; Kqueen, C.Y.; Nishibuchi, M.; Nakaguchi, Y.; Malakar, P.K. Listeria Monocytogenes in Raw Salad Vegetables Sold at Retail Level in Malaysia. Food Control 2010, 21, 774–778. [Google Scholar] [CrossRef]
- Calvo, T. Leafy Greens with Listeria Sold at Major Supermarkets: Consumer Reports Found the Bacteria in Prewashed and Unbagged Products. Available online: https://www.consumerreports.org/food-safety/leafy-greens-with-listeria-sold-at-major-supermarkets/ (accessed on 27 July 2019).
- Sharma, M.; Handy, E.T.; East, C.L.; Kim, S.; Jiang, C.; Callahan, M.T.; Allard, S.M.; Micallef, S.; Craighead, S.; Anderson-Coughlin, B.; et al. Prevalence of Salmonella and Listeria Monocytogenes in Non-Traditional Irrigation Waters in the Mid-Atlantic United States Is Affected by Water Type, Season, and Recovery Method. PLoS ONE 2020, 15, e0229365. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.H.; Vogel, B.F.; Gram, L. Prevalence and Survival of Listeria Monocytogenes in Danish Aquatic and Fish-Processing Environments. J. Food Prot. 2006, 69, 2113–2122. [Google Scholar] [CrossRef] [PubMed]
- NicAogáin, K.; O’Byrne, C.P. The Role of Stress and Stress Adaptations in Determining the Fate of the Bacterial Pathogen Listeria Monocytogenes in the Food Chain. Front. Microbiol. 2016, 7, 1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acheamfour, C.L.; Parveen, S.; Hashem, F.; Sharma, M.; Gerdes, M.E.; May, E.B.; Rogers, K.; Haymaker, J.; Duncan, R.; Foust, D.; et al. Levels of Salmonella Enterica and Listeria Monocytogenes in Alternative Irrigation Water Vary Based on Water Source on the Eastern Shore of Maryland. Microbiol. Spectr. 2021, 9, e00669-21. [Google Scholar] [CrossRef] [PubMed]
- Raschle, S.; Stephan, R.; Stevens, M.J.A.; Cernela, N.; Zurfluh, K.; Muchaamba, F.; Nüesch-Inderbinen, M. Environmental Dissemination of Pathogenic Listeria Monocytogenes in Flowing Surface Waters in Switzerland. Sci. Rep. 2021, 11, 9066. [Google Scholar] [CrossRef]
- Belias, A.; Strawn, L.K.; Wiedmann, M.; Weller, D. Small Produce Farm Environments Can Harbor Diverse Listeria Monocytogenes and Listeria Spp. Populations. J. Food Prot. 2021, 84, 113–121. [Google Scholar] [CrossRef]
- Budzinska, K.; Wronski, G.; Szejniuk, B. Survival Time of Bacteria Listeria Monocytogenes in Water Environment and Sewage. Pol. J. Environ. Stud. 2012, 21, 31–37. [Google Scholar]
- McLaughlin, H.P.; Casey, P.G.; Cotter, J.; Gahan, C.G.M.; Hill, C. Factors Affecting Survival of Listeria Monocytogenes and Listeria Innocua in Soil Samples. Arch. Microbiol. 2011, 193, 775–785. [Google Scholar] [CrossRef]
- Al-ghazali, M.R.; Al-azawi, S.K. Detection and Enumeration of Listeria Monocytogenes in a Sewage Treatment Plant in Iraq. J. Appl. Bacteriol. 1986, 60, 251–254. [Google Scholar] [CrossRef]
- Moreno, Y.; Ballesteros, L.; García-Hernández, J.; Santiago, P.; González, A.; Ferrús, M.A. Specific Detection of Viable Listeria Monocytogenes in Spanish Wastewater Treatment Plants by Fluorescent In Situ Hybridization and PCR. Water Res. 2011, 45, 4634–4640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odjadjare, E.E.O.; Obi, L.C.; Okoh, A.I. Municipal Wastewater Effluents as a Source of Listerial Pathogens in the Aquatic Milieu of the Eastern Cape Province of South Africa: A Concern of Public Health Importance. Int. J. Environ. Res. Public Health 2010, 7, 2376–2394. [Google Scholar] [CrossRef] [PubMed]
- Beneduce, L.; Gatta, G.; Bevilacqua, A.; Libutti, A.; Tarantino, E.; Bellucci, M.; Troiano, E.; Spano, G. Impact of the Reusing of Food Manufacturing Wastewater for Irrigation in a Closed System on the Microbiological Quality of the Food Crops. Int. J. Food Microbiol. 2017, 260, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Shearer, A.E.H.; LeStrange, K.; Castaneda SALDAÑA, R.; Kniel, K.E. Transfer of Pathogens from Cantaloupe Rind to Preparation Surfaces and Edible Tissue as a Function of Cutting Method. J. Food Prot. 2016, 79, 764–770. [Google Scholar] [CrossRef] [Green Version]
- U.S. Environmental Protection. Agency National Polluntant Discharge Elimination System (NPDES): Municipal Wastewater. Available online: https://www.epa.gov/npdes/municipal-wastewater (accessed on 2 September 2019).
- U.S. Environmental Protection. Agency Approved CWA Microbiological Test Methods. Available online: https://www.epa.gov/cwa-methods/approved-cwa-microbiological-test-methods (accessed on 19 August 2018).
- Guévremont, E.; Lamoureux, L.; Généreux, M.; Côté, C. Irrigation Water Sources and Time Intervals as Variables on the Presence of Campylobacter Spp. and Listeria Monocytogenes on Romaine Lettuce Grown in Muck Soil. J. Food Prot. 2017, 80, 1182–1187. [Google Scholar] [CrossRef]
- Oliveira, M.; Usall, J.; Viñas, I.; Solsona, C.; Abadias, M. Transfer of Listeria Innocua from Contaminated Compost and Irrigation Water to Lettuce Leaves. Food Microbiol. 2011, 28, 590–596. [Google Scholar] [CrossRef]
- Kühbacher, A.; Cossart, P.; Pizarro-Cerdá, J. Internalization Assays for Listeria Monocytogenes. In Listeria Monocytogenes; Humana Press: New York, NY, USA, 2014; pp. 167–178. [Google Scholar]
- Portnoy, D.A.; Jacks, P.S.; Hinrichs, D.J. Role of Hemolysin for the Intracellular Growth of Listeria Monocytogenes. J. Exp. Med. 1988, 167, 1459–1471. [Google Scholar] [CrossRef] [Green Version]
- Xerry, J.; Gallimore, C.I.; Iturriza-Gomara, M.; Gray, J. Tracking the transmission routes of genogroup II noroviruses in suspected food-borne or environmental outbreaks of gastroenteritis through sequence analysis of the P2 domain. J. Med. Virol. 2009, 1304, 1298–1304. [Google Scholar] [CrossRef]
- Food Microbe Tracker Food Microbe Tracker: The Cornell Food Safety Laboratory Bacterial Strains Www Database Project. Available online: www.pathogentracker.net (accessed on 2 September 2019).
- Iwu, C.D.; Okoh, A.I. Characterization of Antibiogram Fingerprints in Listeria Monocytogenes Recovered from Irrigation Water and Agricultural Soil Samples. PLoS ONE 2020, 15, e0228956. [Google Scholar] [CrossRef] [Green Version]
| Country | Positive/Total Samples (% Positive) | Type of Water | Factors Contributing to Presence | Reference |
|---|---|---|---|---|
| USA (California) | 605/1405 (43) | Lake, stream, river, pond | Point-source, roaming animals, high run-off from heavy rains | [10] |
| USA (New York) | 48/174 (28) | Surface and engineered | Farm, season, sample type, temperature | [12] |
| USA (New York) | 22/74 (30) | Irrigation (engineered, pond, river) and non-irrigation (pond, ditch, river) | Water source (non-irrigation vs. irrigation) | [11] |
| Canada (Ontario) | 32/134 (10) | Surface (river) | Proximity to upstream dairy farm, degree of crop land | [42] |
| Germany | 24/36 (67) | Creek and pond | Area rich in agriculture and plant life | [37] |
| Austria | 0/68 (0) | River and pond | Proximity to agricultural lands, urban environments | [38] |
| South Africa | 19/36 (53) | Irrigation canal and river | High chemical oxygen demand detected | [8] |
| India (Varanasi) | 8/100 (8) | River Water | Proximity to large human population | [47] |
| Canada (Nova Scotia) | 56/329 (17) | Rural and urban watersheds (river and lake) | Rural agricultural watersheds | [24] |
| USA (New York) | 10/33 (30) | Irrigation (well, pond) and non-irrigation (ditch creek) | Sources of water, more likely in non-irrigation sources | [20] |
| USA (Mid-Atlantic) | 53/171 (31) | Pond, non-tidal fresh, tidal fresh, tidal brackish, reclaimed | Water source (environmental vs. reclaimed) | [74,77] |
| Switzerland | 25/191 (13) | River, stream, inland canal | Agricultural area and dense human populations | [78] |
| Denmark | 2/26 (8) | Stream, freshwater fish farm, seawater fish farm, municipal water | Prevalence increased with degree of human activity | [75] |
| USA (New York) | 86/209 (41%) | Pond, stream, and wildlife fecal samples | Small farm with wildlife intrusion | [79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gartley, S.; Anderson-Coughlin, B.; Sharma, M.; Kniel, K.E. Listeria monocytogenes in Irrigation Water: An Assessment of Outbreaks, Sources, Prevalence, and Persistence. Microorganisms 2022, 10, 1319. https://doi.org/10.3390/microorganisms10071319
Gartley S, Anderson-Coughlin B, Sharma M, Kniel KE. Listeria monocytogenes in Irrigation Water: An Assessment of Outbreaks, Sources, Prevalence, and Persistence. Microorganisms. 2022; 10(7):1319. https://doi.org/10.3390/microorganisms10071319
Chicago/Turabian StyleGartley, Samantha, Brienna Anderson-Coughlin, Manan Sharma, and Kalmia E. Kniel. 2022. "Listeria monocytogenes in Irrigation Water: An Assessment of Outbreaks, Sources, Prevalence, and Persistence" Microorganisms 10, no. 7: 1319. https://doi.org/10.3390/microorganisms10071319
APA StyleGartley, S., Anderson-Coughlin, B., Sharma, M., & Kniel, K. E. (2022). Listeria monocytogenes in Irrigation Water: An Assessment of Outbreaks, Sources, Prevalence, and Persistence. Microorganisms, 10(7), 1319. https://doi.org/10.3390/microorganisms10071319