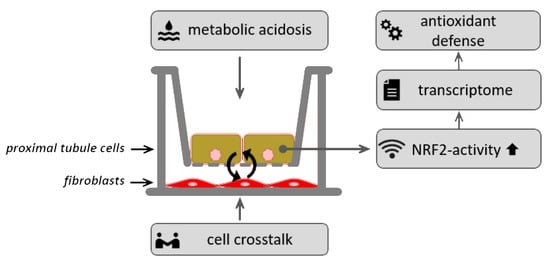
Acidosis Activates the Nrf2 Pathway in Renal Proximal Tubule-Derived Cells through a Crosstalk with Renal Fibroblasts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Experimental Setup
2.3. RNA Sample Preparation
2.4. RNA Sequencing
2.5. Differential Expression Analysis and Functional Analysis
2.6. Quantitative PCR
2.7. Cell Fractionation
2.8. Western Blot
2.9. ARE-Luciferase Assay
2.10. Data Analysis
3. Results
3.1. Impact of Extracellular Acidosis on General Gene Expression
3.2. Genes Regulated by Extracellular Acidosis in Both Cell Lines Are Enriched in the Nrf2 Pathway
3.3. Impact of Extracellular Acidosis on Nrf2 Expression
3.4. Impact of Extracellular Acidosis on Nrf2 Phosphorylation and Activation
3.5. Acidosis Leads to Enhanced Expression of the Canonical Nrf2 Target Gene TKT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Neuen, B.L.; Chadban, S.J.; Demaio, A.R.; Johnson, D.W.; Perkovic, V. Chronic kidney disease and the global NCDs agenda. BMJ Glob. Health 2017, 2, e000380. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, R.L. The proximal tubule is the primary target of injury and progression of kidney disease: Role of the glomerulotubular junction. Am. J. Physiol. Renal Physiol. 2016, 311, F145–F161. [Google Scholar] [CrossRef]
- Schnaper, H.W. The Tubulointerstitial Pathophysiology of Progressive Kidney Disease. Adv. Chronic Kidney Dis. 2017, 24, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Kellum, J.A.; Song, M.; Li, J. Science review: Extracellular acidosis and the immune response: Clinical and physiologic implications. Crit. Care 2004, 8, 331–336. [Google Scholar] [CrossRef]
- Wesson, D.E.; Buysse, J.M.; Bushinsky, D.A. Mechanisms of Metabolic Acidosis-Induced Kidney Injury in Chronic Kidney Disease. J. Am. Soc. Nephrol. 2020, 31, 469–482. [Google Scholar] [CrossRef]
- Kuramochi, G.; Gekle, M.; Silbernagl, S. Derangement of pH homeostasis in renal papilla: Ochratoxin A increases pH in vasa recta blood. Nephron 1997, 76, 472–476. [Google Scholar] [CrossRef]
- Kuramochi, G.; Gekle, M.; Silbernagl, S. Ochratoxin A disturbs pH homeostasis in the kidney: Increases in pH and HCO3− in the tubule and vasa recta. Pflügers Arch. 1997, 434, 392–397. [Google Scholar] [CrossRef]
- Kuramochi, G.; Kersting, U.; Dantzler, W.H.; Silbernagl, S. Changes in the coutercurrent system in the renal papilla: Diuresis increases pH and HCO3− gradients between collecting duct and vasa recta. Pflügers Arch. 1996, 432, 1062–1068. [Google Scholar] [CrossRef]
- Magno, A.L.; Herat, L.Y.; Carnagarin, R.; Schlaich, M.P.; Matthews, V.B. Current Knowledge of IL-6 Cytokine Family Members in Acute and Chronic Kidney Disease. Biomedicines 2019, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovisa, S.; LeBleu, V.S.; Tampe, B.; Sugimoto, H.; Vadnagara, K.; Carstens, J.L.; Wu, C.C.; Hagos, Y.; Burckhardt, B.C.; Pentcheva-Hoang, T.; et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat. Med. 2015, 21, 998–1009. [Google Scholar] [CrossRef]
- Ferenbach, D.A.; Bonventre, J.V. Kidney tubules: Intertubular, vascular, and glomerular cross-talk. Curr. Opin. Nephrol. Hypertens. 2016, 25, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Bülow, R.D.; Boor, P. Extracellular Matrix in Kidney Fibrosis: More than Just a Scaffold. J. Histochem. Cytochem. 2019, 67, 643–661. [Google Scholar] [CrossRef]
- Zhou, D.; Liu, Y. Renal fibrosis in 2015: Understanding the mechanisms of kidney fibrosis. Nat. Rev. Nephrol. 2016, 12, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Prunotto, M.; Budd, D.C.; Gabbiani, G.; Meier, M.; Formentini, I.; Hartmann, G.; Pomposiello, S.; Moll, S. Epithelial-mesenchymal crosstalk alteration in kidney fibrosis. J. Pathol. 2012, 228, 131–147. [Google Scholar] [CrossRef]
- Schulz, M.C.; Voß, L.; Schwerdt, G.; Gekle, M. Epithelial-Fibroblast Crosstalk Protects against Acidosis-Induced Inflammatory and Fibrotic Alterations. Biomedicines 2022, 10, 681. [Google Scholar] [CrossRef]
- Noel, S.; Hamad, A.R.; Rabb, H. Reviving the promise of transcription factor Nrf2-based therapeutics for kidney diseases. Kidney Int. 2015, 88, 1217–1218. [Google Scholar] [CrossRef]
- Nezu, M.; Souma, T.; Yu, L.; Suzuki, T.; Saigusa, D.; Ito, S.; Suzuki, N.; Yamamoto, M. Transcription factor Nrf2 hyperactivation in early-phase renal ischemia-reperfusion injury prevents tubular damage progression. Kidney Int. 2017, 91, 387–401. [Google Scholar] [CrossRef]
- Shelton, L.M.; Lister, A.; Walsh, J.; Jenkins, R.E.; Wong, M.H.; Rowe, C.; Ricci, E.; Ressel, L.; Fang, Y.; Demougin, P.; et al. Integrated transcriptomic and proteomic analyses uncover regulatory roles of Nrf2 in the kidney. Kidney Int. 2015, 88, 1261–1273. [Google Scholar] [CrossRef]
- de la Vega, M.R.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Warabi, E.; Mann, G.E. Mechanisms underlying Nrf2 nuclear translocation by non-lethal levels of hydrogen peroxide: p38 MAPK-dependent neutral sphingomyelinase2 membrane trafficking and ceramide/PKC/CK2 signaling. Free Radic. Biol. Med. 2022, 191, 191–202. [Google Scholar] [CrossRef]
- Kim, D.E.; Byeon, H.E.; Kim, D.H.; Kim, S.G.; Yim, H. Plk2-mediated phosphorylation and translocalization of Nrf2 activates anti-inflammation through p53/Plk2/p21(cip1) signaling in acute kidney injury. Cell Biol. Toxicol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Bellezza, I.; Giambanco, I.; Minelli, A.; Donato, R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Lister, A.; Bourgeois, S.; Imenez Silva, P.H.; Rubio-Aliaga, I.; Marbet, P.; Walsh, J.; Shelton, L.M.; Keller, B.; Verrey, F.; Devuyst, O.; et al. NRF2 regulates the glutamine transporter Slc38a3 (SNAT3) in kidney in response to metabolic acidosis. Sci. Rep. 2018, 8, 5629. [Google Scholar] [CrossRef]
- Romero-Garcia, S.; Prado-Garcia, H.; Valencia-Camargo, A.D.; Alvarez-Pulido, A. Lactic Acidosis Promotes Mitochondrial Biogenesis in Lung Adenocarcinoma Cells, Supporting Proliferation Under Normoxia or Survival Under Hypoxia. Front. Oncol. 2019, 9, 1053. [Google Scholar] [CrossRef]
- Katsu-Jimenez, Y.; Vazquez-Calvo, C.; Maffezzini, C.; Halldin, M.; Peng, X.; Freyer, C.; Wredenberg, A.; GimÃnez-Cassina, A.; Wedell, A.; Arner, E.S.J. Absence of TXNIP in Humans Leads to Lactic Acidosis and Low Serum Methionine Linked to Deficient Respiration on Pyruvate. Diabetes 2019, 68, 709–723. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guyguen, L.; Coppe, J.Y.; Dillies, M.A.s. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Ruhs, S.; Strätz, N.; Quarch, K.; Masch, A.; Schutkowski, M.; Gekle, M.; Grossmann, C. Modulation of transcriptional mineralocorticoid receptor activity by casein kinase 2. Sci. Rep. 2017, 7, 15340. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Li, Z.; Leffler, N.R.; Asch, A.S.; Chi, J.T.; Yang, L.V. Acidosis activation of the proton-sensing GPR4 receptor stimulates vascular endothelial cell inflammatory responses revealed by transcriptome analysis. PLoS ONE 2013, 8, e61991. [Google Scholar] [CrossRef]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. USA 2000, 97, 12475–12480. [Google Scholar] [CrossRef]
- Hao, S.; Meng, Q.; Sun, H.; Li, Y.; Li, Y.; Gu, L.; Liu, B.; Zhang, Y.; Zhou, H.; Xu, Z.; et al. The role of transketolase in human cancer progression and therapy. Biomed. Pharmacother. 2022, 154, 113607. [Google Scholar] [CrossRef]
- Xu, I.M.; Lai, R.K.; Lin, S.H.; Tse, A.P.; Chiu, D.K.; Koh, H.Y.; Law, C.T.; Wong, C.M.; Cai, Z.; Wong, C.C.; et al. Transketolase counteracts oxidative stress to drive cancer development. Proc. Natl. Acad. Sci. USA 2016, 113, E725–E734. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Target | Company | Order Number | Host | Dilution |
|---|---|---|---|---|
| Antirabbit IgG HRP | Cell Signaling, Danvers, MA, USA | 7074 | Goat | 1:1000 |
| NRF2 (C-20) | Santa Cruz Biotechnology, Inc., TX, USA | sc-722 | Rabbit | 1:1000 |
| Phospho-NRF2 (Ser40) | Thermo Fisher Scientific GmbH, Dreieich, Germany | PA5-67520 | Rabbit | 1:500 |
| Transketolase | Cell Signaling, Danvers, MA, USA | E7O4M | Rabbit | 1:2000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schulz, M.-C.; Dubourg, V.; Nolze, A.; Kopf, M.; Schwerdt, G.; Gekle, M. Acidosis Activates the Nrf2 Pathway in Renal Proximal Tubule-Derived Cells through a Crosstalk with Renal Fibroblasts. Antioxidants 2023, 12, 412. https://doi.org/10.3390/antiox12020412
Schulz M-C, Dubourg V, Nolze A, Kopf M, Schwerdt G, Gekle M. Acidosis Activates the Nrf2 Pathway in Renal Proximal Tubule-Derived Cells through a Crosstalk with Renal Fibroblasts. Antioxidants. 2023; 12(2):412. https://doi.org/10.3390/antiox12020412
Chicago/Turabian StyleSchulz, Marie-Christin, Virginie Dubourg, Alexander Nolze, Michael Kopf, Gerald Schwerdt, and Michael Gekle. 2023. "Acidosis Activates the Nrf2 Pathway in Renal Proximal Tubule-Derived Cells through a Crosstalk with Renal Fibroblasts" Antioxidants 12, no. 2: 412. https://doi.org/10.3390/antiox12020412
APA StyleSchulz, M. -C., Dubourg, V., Nolze, A., Kopf, M., Schwerdt, G., & Gekle, M. (2023). Acidosis Activates the Nrf2 Pathway in Renal Proximal Tubule-Derived Cells through a Crosstalk with Renal Fibroblasts. Antioxidants, 12(2), 412. https://doi.org/10.3390/antiox12020412