1. Introduction
The electrical and thermal energy production processes that use non-renewable resources (i.e., fossil fuels; oil, and coal) are becoming less attractive globally. Even though such resources are rich in energy and relatively inexpensive to process, they are limited in supply and will soon be depleted. In addition, the utilization of fossil fuels emits additional greenhouse gases into the atmosphere, which has instigated climate change [
1]. Hence, a large number of research bodies have aligned to overcome such an increasing universal concern. One of the most promising and attractive alternative solutions is the use of biogas derived from wastes or renewable feedstock [
2,
3].
Biogas, a mixture consisting chiefly of methane (CH
4) and carbon dioxide (CO
2), is the end-product of anaerobic digestion of organic matters (e.g., agricultural residues, livestock manure, food waste, sewage sludge, etc.) [
4,
5,
6,
7,
8]. Anaerobic digestion is a complex multi-step process that is carried out by a consortium of different microbial species known as anaerobes. Uniquely, they do not need molecular oxygen for their metabolism and growth [
9]. The key steps of the anaerobic digestion process, together with the possible applications of biogas, and its adverse environmental impacts are outlined in
Figure 1.
The increasing global interest in biogas power plant establishment via anaerobic digestion of various organic matters has resulted in attempts to develop numerous mathematical models to predict and suggest optimal operations. Hill [
15] developed a model to describe the digestion of animal wastes, assuming that the main five bacterial groups involved in the overall digestion process (acidogenic bacteria, hydrogenotrophic bacteria, homoacetogenic bacteria, acetoclastic bacteria, and H
2 utilizing methane bacteria) are inhibited by a high concentration of fatty acids (FAs). Mosey [
16] proposed a model consisting of four reactions (one acidogenic reaction, one acetogenic reaction, and two methanogenic reactions), which also takes into account the role of H
2. According to this model, in case of a sudden rise in the organic loading rate, an accumulation of volatile fatty acids (VFAs) is likely to occur; this results in a decrease in pH that inhibits H
2 utilizing methanogenic bacteria. In other words, H
2 partial pressure is increased, which leads to further accumulation of propionic/butyric acid (CH
4 generation is stopped when pH drops below 5.5). Based on Mosey’s model, Pullammanappallil et al. [
17] introduced a model taking into account the gas phase, and acetoclastic inhibition by undissociated FAs. Angelidaki et al. [
18] presented a model considering hydrolysis, acidogenesis, acetogenesis, and methanogenesis, which is suitable to describe the behavior of anaerobic digesters fed with manures. This model was developed by incorporating some assumptions as follows: (i) methanogenesis is inhibited by free NH
3, (ii) acetogenesis is inhibited by acetic acid, (iii) acidogenesis is inhibited by total VFAs, and (iv) the degree of NH
3 ionization, the maximum specific growth rate of bacteria are pH and temperature dependent.
In all the above-mentioned models, organic material was taken into account as a whole; in other words, they are incapable of dealing with complex feed composition. In this regard, the International Water Association (IWA) task group for mathematical modeling of the anaerobic digestion process developed a model known as Anaerobic Digester Model No 1 (more often abbreviated as ADM1), that takes the complex organic substrates into account [
19].
Although the kinetic-based mathematical models for describing the anaerobic digestion process can help engineers and asset managers to better plan the management of the biogas plants, it is often criticized that most of them are inherently too complex due to a large number of stoichiometric coefficients and parameters reflecting the kinetic properties of the enzymes and microorganisms that govern the physicochemical and biochemical reactions through anaerobic digestion processes [
20]. In addition, these models typically involve physicochemical equilibrium expressions and differential mass balance equations for components in the liquid phase (substrates for acidogenic/acetogenic/methanogenic organisms and their corresponding microbial masses) and in the gas phase (e.g., CH
4 and CO
2). Hence, these models are often complicated to solve, and many simplifying assumptions must be made to reduce their complexity. However, incorporating simplifying assumptions into the models may not hold in practice. Fedailaine et al. [
21] modeled the biokinetics of the anaerobic digestion process involving eight simplifying assumptions, which inevitably limited the application of this model to full-scale anaerobic digesters. In addition, applying assumptions to the models lowers the precision of the models; in other words, an under- or over-estimation of the response of the models will likely occur. For these reasons, developing a simple yet highly predictive model to estimate biogas production from the anaerobic digestion process is highly desired. As such, a different branch of models, called artificial intelligence (AI)-based models (more often known as easy-to-use black-box models) may be recruited. These models have advantages over complex mathematical models because they are constructed on a measured dataset (i.e., input–output data pairs for a given system) without requiring complicated kinetic relationships between the input variables and the corresponding outputs [
22,
23]. In addition, the AI modeling approach is proven as a robust tool with high generalization power. Holubar et al. [
24] used an artificial neural network (ANN) to model an anaerobic digester fed with a mixture of primary (raw) sludge and surplus activated sludge originating from a local municipal wastewater treatment plant. The results showed that ANN is a suitable tool for modeling such a process. Cakmakci [
25] applied an adaptive neuro-fuzzy inference system (ANFIS) to predict methane yield in an anaerobic digester fed with pre-thickened raw sludge. According to the findings, there was good agreement between the measured and predicted values. Kusiak and Wei [
26] developed several predictive models through data mining algorithms to predict methane production from the anaerobic digesters in the Des Moines Wastewater Reclamation Facility. The results showed that the model built by the ANFIS algorithm offered excellent predictive accuracy with a coefficient of determination (
R2) of 0.99, and a percentage error of 0.08. Nair et al. [
27] used ANN to evaluate the effects of the types of substrates (such as food/vegetable waste and yard trimming), and organic loading rate on CH
4 production. The training and validation
R2 values were greater than 0.88, indicating that the model’s learning and generalization power were satisfactory. Dach et al. [
28] reported that ANN can be considered an appropriate tool to estimate CH
4 from anaerobic digestion of slurry from animal waste and agricultural residues. Tan et al. [
29] compared the performance of ANFIS and the ADM1 to predict biogas production from the anaerobic digestion of palm oil mill effluent under thermophilic conditions. The authors reported that ANFIS yielded higher predictive accuracy compared with the results obtained using the ADM1. In another study conducted by Beltramo et al. [
30], an ANN model was constructed to predict the biogas production rate from a mesophilic anaerobic digester fed with a mixture of maize, grass silages, and pig/cattle manure. The authors conclude that the ANN modeling approach can be considered a promising alternative to ADM1.
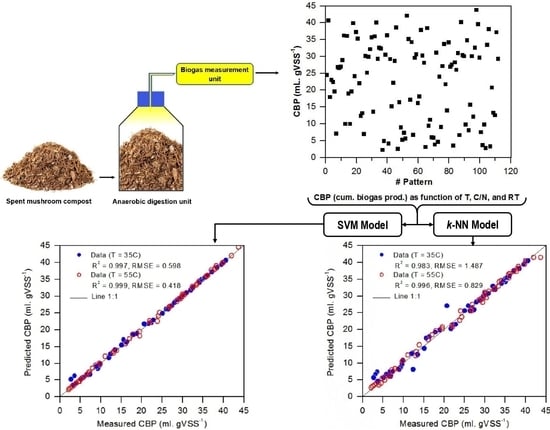
This study aimed to develop, validate, and test two different predictive models based on the AI modeling approach, including k-nearest neighbors and support vector machine (referred to hereafter as k-NN and SVM, respectively) to predict biogas production from anaerobic digestion of spent mushroom compost (SMC). The independent variables involved include temperature, carbon-to-nitrogen ratio (C/N), and retention time (RT). SMC is a bulky residue from mushroom farms, and the waste generated by the mushroom processing industry. It is an ideal source of general nutrients (e.g., nitrogen and phosphorus) and is rich in organic matter that can be used for producing biogas. It is worth mentioning that the nutritional value and the content of organic matter of SMC depend on the types of cultivated mushroom species.
The predictive performance of these models was separately investigated and eventually compared with each other and with the ANN, ANFIS, and logistic models developed by Najafi and Faizollahzadeh Ardabili [
31] by means of two statistical indices, including
R2, and root mean squared error (
RMSE). To the best of the authors’ knowledge, the application of
k-NN and SVM modeling approaches to predict biogas production from
SMC has never been exploited.