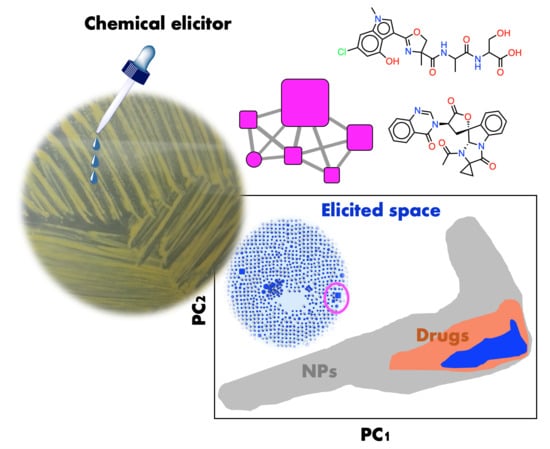
Chemical Elicitors Induce Rare Bioactive Secondary Metabolites in Deep-Sea Bacteria under Laboratory Conditions
Abstract
:1. Introduction
2. Results
2.1. Construction of the Elicited Pre-Fractionated Bacterial (EPfB) Chemical Library
2.1.1. Collection of Deep-Sea Bacteria
2.1.2. Implementation of a New Method for Testing Growth Conditions and Chemical Elicitors for Bacterial Natural Product Library Construction
2.1.3. Preparation of the EPfB Chemical Library
2.2. Chemical Composition of the EPfB Chemical Library
2.2.1. Global Analysis of the Bacterial Metabolites Contained in the EPfB Library
2.2.2. Compound Novelty Introduced by Eliciting Bacterial Cultures
2.2.3. Chemical Diversity of the EPfB Library
2.2.4. Biological Relevance of the Library
2.2.5. Chemical Redundancy in the Relevant Biological Space
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bruder, M.; Polo, G.; Trivella, D.B. Natural allosteric modulators and their biological targets: Molecular signatures and mechanisms. Nat. Prod. Rep. 2020, 37, 488–514. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Ghoul, M.; Mitri, S. The Ecology and Evolution of Microbial Competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Macheboeuf, P.; Contreras-Martel, C.; Job, V.; Dideberg, O.; Dessen, A. Penicillin Binding Proteins: Key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006, 30, 673–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawara, K.; Hatori, M.; Nishiyama, Y.; Tomita, K.; Kamei, H.; Konishi, M.; Oki, T. Eponemycin, a new antibiotic active against B16 melanoma. I. Production, isolation, structure and biological activity. J. Antibiot. (Tokyo) 1990, 43, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Groll, M.; Kim, K.B.; Kairies, N.; Huber, R.; Crews, C.M. Crystal Structure of Epoxomicin:20S Proteasome Reveals a Molecular Basis for Selectivity of α′,β′-Epoxyketone Proteasome Inhibitors. J. Am. Chem. Soc. 2000, 122, 1237–1238. [Google Scholar] [CrossRef]
- Zhang, X.; Linder, S.; Bazzaro, M. Drug Development Targeting the Ubiquitin–Proteasome System (UPS) for the Treatment of Human Cancers. Cancers 2020, 12, 902. [Google Scholar] [CrossRef] [Green Version]
- Ciechanover, A. The ubiquitin-proteasome proteolytic pathway. Cell 1994, 79, 13–21. [Google Scholar] [CrossRef]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A Highly Cytotoxic Proteasome Inhibitor from a Novel Microbial Source, a Marine Bacterium of the New Genus Salinospora. Angew. Chem. Int. Ed. 2003, 42, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Tundo, G.R.; Sbardella, D.; Santoro, A.M.; Coletta, A.; Oddone, F.; Grasso, G.; Milardi, D.; Lacal, P.; Marini, S.; Purrello, P.; et al. The proteasome as a druggable target with multiple therapeutic potentialities: Cutting and non-cutting edges. Pharmacol. Ther. 2020, 213, 107579. [Google Scholar] [CrossRef]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Kale, A.J.; McGlinchey, R.P.; Lechner, A.; Moore, B.S. Bacterial Self-Resistance to the Natural Proteasome Inhibitor Salinosporamide A. ACS Chem. Biol. 2011, 6, 1257–1264. [Google Scholar] [CrossRef] [Green Version]
- Shank, E.A.; Kolter, R. New developments in microbial interspecies signaling. Curr. Opin. Microbiol. 2009, 12, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Pettit, R.K. Small-molecule elicitation of microbial secondary metabolites. Microb. Biotechnol. 2011, 4, 471–478. [Google Scholar] [CrossRef] [Green Version]
- Trivella, D.B.B.; de Felicio, R. The Tripod for Bacterial Natural Product Discovery: Genome Mining, Silent Pathway Induction, and Mass Spectrometry-Based Molecular Networking. mSystems 2018, 3, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seyedsayamdost, M.R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. USA 2014, 111, 7266–7271. [Google Scholar] [CrossRef] [Green Version]
- Tomm, H.A.; Ucciferri, L.; Ross, A.C. Advances in microbial culturing conditions to activate silent biosynthetic gene clusters for novel metabolite production. J. Ind. Microbiol. Biotechnol. 2019, 46, 1381–1400. [Google Scholar] [CrossRef]
- Lynn, K.-S.; Cheng, M.-L.; Chen, Y.-R.; Hsu, C.; Chen, A.; Lih, T.M.; Chang, H.-Y.; Huang, C.; Shiao, M.-S.; Pan, W.-H.; et al. Metabolite Identification for Mass Spectrometry-Based Metabolomics Using Multiple Types of Correlated Ion Information. Anal. Chem. 2015, 87, 2143–2151. [Google Scholar] [CrossRef]
- Riquelme, G.; Zabalegui, N.; Marchi, P.; Jones, C.M.; Monge, M.E. A Python-Based Pipeline for Preprocessing LC–MS Data for Untargeted Metabolomics Workflows. Metabolites 2020, 10, 416. [Google Scholar] [CrossRef] [PubMed]
- Stricker, T.; Bonner, R.; Lisaek, F.; Hopfgartner, G. Adduct annotation in liquid chromatography/high-resolution mass spectrometry to enhance compound identification. Anal. Bioanal. Chem. 2020, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.-F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K. Insights into microbial cryptic gene activation and strain improvement: Principle, application and technical aspects. J. Antibiot. 2017, 70, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, K.; Takagi, M.; Shin-ya, K. Tetrapeptides Possessing a Unique Skeleton, JBIR-34 and JBIR-35, Isolated from a Sponge-Derived Actinomycete, Streptomyces sp. Sp080513GE-23. J. Nat. Prod. 2010, 73, 226–228. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Debbab, A.; Mándi, A.; Wray, V.; Schulz, B.; Müller, W.E.G.; Kassack, M.; Lin, W.; Kurtán, T.; Proksch, P.; et al. Alkaloids from the Sponge-Associated Fungus Aspergillus sp. Eur. J. Org. Chem. 2013, 2013, 894–906. [Google Scholar] [CrossRef]
- Li, X.-J.; Zhang, Q.; Zhang, A.-L.; Gao, J.-M. Metabolites from Aspergillus fumigatus, an Endophytic Fungus Associated with Melia azedarach, and Their Antifungal, Antifeedant, and Toxic Activities. J. Agric. Food Chem. 2012, 60, 3424–3431. [Google Scholar] [CrossRef]
- Peng, J.; Lin, T.; Wang, W.; Xin, Z.; Zhu, T.; Gu, Q.; Li, D. Antiviral Alkaloids Produced by the Mangrove-Derived Fungus Cladosporium sp. PJX-41. J. Nat. Prod. 2013, 76, 1133–1140. [Google Scholar] [CrossRef]
- Ahammad Uz Zaman, K.; Hu, Z.; Wu, X.; Cao, S. Tryptoquivalines W and X, two new compounds from a Hawaiian fungal strain and their biological activities. Tetrahedron Lett. 2020, 61, 151730. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, S.-X.; Ju, J.; Tang, G.; Liu, T.; Shen, B. Characterization of the lnmKLM Genes Unveiling Key Intermediates for β-Alkylation in Leinamycin Biosynthesis. Org. Lett. 2011, 13, 498–501. [Google Scholar] [CrossRef] [Green Version]
- Nogawa, T.; Okano, A.; Takahashi, S.; Uramoto, M.; Konno, H.; Saito, T.; Osada, H. Verticilactam, a New Macrolactam Isolated from a Microbial Metabolite Fraction Library. Org. Lett. 2010, 12, 4564–4567. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.-J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maneerat, S.; Nitoda, T.; Kanzaki, H.; Kawai, F. Bile acids are new products of a marine bacterium, Myroides sp. strain SM1. Appl. Microbiol. Biotechnol. 2005, 67, 679–683. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.S.; Kim, J.; Kang, S.-J.; Yoon, J.-H.; Kim, W.G.; Lee, C.H. Biosynthesis of bile acids in a variety of marine bacterial taxa. J. Microbiol. Biotechnol. 2007, 17, 403–407. [Google Scholar]
- Choudhary, A.; Naughton, L.M.; Dobson, A.D.W.; Rai, D.K. Metabolites from Aspergillus fumigatus, an Endophytic Fungus Associated with Melia azedarach, and their lites produced by Pseudovibrio sp. W64, a marine sponge derived bacterium isolated from Irish waters. Rapid Commun. Mass Spectrom. 2018, 32, 1737–1745. [Google Scholar] [CrossRef] [PubMed]
- Dobson, T.E.; Maxwell, A.R.; Ramsubhag, A. Antimicrobial cholic acid derivatives from the Pitch Lake bacterium Bacillus amyloliquefaciens UWI-W23. Steroids 2018, 135, 50–53. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Willighagen, E.L.; Mayfield, J.W.; Alvarsson, J.; Berg, A.; Carlsson, L.; Jeliazkova, N.; Kuhn, S.; Pluskal, T.; Rojas-Chertó, M.; Spjuth, O.; et al. The Chemistry Development Kit (CDK) v2.0: Atom typing, depiction, molecular formulas, and substructure searching. J. Cheminform. 2017, 9, 33. [Google Scholar] [CrossRef] [Green Version]
- Dobson, C.M. Chemical space and biology. Nature 2004, 432, 824–828. [Google Scholar] [CrossRef]
- Auvin, C.; Lezenven, F.; Blond, A.; Augeven-Bour, I.; Pousset, J.-L.; Bodo, B.; Camara, J. Mucronine J, a 14-Membered Cyclopeptide Alkaloid from Zizyphus mucronata. J. Nat. Prod. 1996, 59, 676–678. [Google Scholar] [CrossRef] [PubMed]
- Tschesche, R.; Elgamal, M.; Miana, G.A.; Eckhardt, G. Alkaloids from rhamnaceae—XXVI: Nummularine -D, -E and -F, new cyclopeptide alkaloids from Zizyphus nummularia. Tetrahedron 1975, 31, 2944–2947. [Google Scholar] [CrossRef]
- Lax, A.R.; Shepherd, H.S. Tentoxin: A Cyclic Tetrapeptide Having Potential Herbicidal Usage. In Biologically Active Natural Products; Cutler, H.G., Ed.; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1988; Volume 380, pp. 24–34. ISBN 0-8412-1556-1. [Google Scholar]
- Rungprom, W.; Siwu, E.R.O.; Lambert, L.K.; Dechsakulwatana, C.; Barden, M.C.; Kokpol, U.; Blanchfield, J.T.; Kita, M.; Garson, M.J. Cyclic tetrapeptides from marine bacteria associated with the seaweed Diginea sp. and the sponge Halisarca ectofibrosa. Tetrahedron 2008, 64, 3147–3152. [Google Scholar] [CrossRef]
- Kobayashi, J.; Kubota, T. The Daphniphyllum alkaloids. Nat. Prod. Rep. 2009, 26, 936. [Google Scholar] [CrossRef]
- Luesch, H.; Williams, P.G.; Yoshida, W.Y.; Moore, R.E.; Paul, V.J. Ulongamides A−F, New β-Amino Acid-Containing Cyclodepsipeptides from Palauan Collections of the Marine Cyanobacterium Lyngbya sp. J. Nat. Prod. 2002, 65, 996–1000. [Google Scholar] [CrossRef]
- Wolfender, J.-L.; Nuzillard, J.-M.; van der Hooft, J.J.J.; Renault, J.-H.; Bertrand, S. Accelerating Metabolite Identification in Natural Product Research: Toward an Ideal Combination of Liquid Chromatography–High-Resolution Tandem Mass Spectrometry and NMR Profiling, in Silico Databases, and Chemometrics. Anal. Chem. 2019, 91, 704–742. [Google Scholar] [CrossRef]
- Baral, B.; Akhgari, A.; Metsä-Ketelä, M. Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178. [Google Scholar] [CrossRef]
- Kobayashi, J.; Cheng, J.; Walchli, M.R.; Nakamura, H.; Hirata, Y.; Sasaki, T.; Ohizumi, Y. Cystodytins A, B, and C, novel tetracyclic aromatic alkaloids with potent antineoplastic activity from the Okinawan tunicate Cystodytes dellechiajei. J. Org. Chem. 1988, 53, 1800–1804. [Google Scholar] [CrossRef]
- Wilson, B.A.P.; Thornburg, C.C.; Henrich, C.J.; Grkovic, T.; O’Keefe, B.R. Creating and screening natural product libraries. Nat. Prod. Rep. 2020, 37, 893–918. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, M.M. Pre-fractionated microbial samples—The second generation natural products library at Wyeth. Molecules 2008, 13, 1406–1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva, M.A.C.; Cavalett, A.; Spinner, A.; Rosa, D.C.; Jasper, R.B.; Quecine, M.C.; Bonatelli, M.L.; Pizzirani-Kleiner, A.; Corção, G.; Lima, A.O.d.S. Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. Springerplus 2013, 2, 127. [Google Scholar] [CrossRef] [Green Version]
- De Souza Lima, A.O.; Cabral, A.; Andreote, F.D.; Cavalett, A.; Pessatti, M.L.; Dini-Andreote, F.; da Silva, M.A.C. Draft Genome Sequence of Bacillus stratosphericus LAMA 585, Isolated from the Atlantic Deep Sea. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [Green Version]
- Grkovic, T.; Akee, R.K.; Thornburg, C.C.; Trinh, S.K.; Britt, J.R.; Harris, M.J.; Evans, J.R.; Kang, U.; Ensel, S.; Henrich, C.J.; et al. National Cancer Institute (NCI) Program for Natural Products Discovery: Rapid Isolation and Identification of Biologically Active Natural Products from the NCI Prefractionated Library. ACS Chem. Biol. 2020, 15, 1104–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waechter, F.; da Silva, G.N.S.; Willig, J.B.; de Oliveira, C.B.; Vieira, B.D.; Trivella, D.B.B.; Zimmer, A.R.; Buffon, A.; Pilger, D.A.; Gnoatto, S.C.B. Design, synthesis and biological evaluation of betulinic acid derivatives as new antitumor agents for leukemia. Anticancer Agents Med. Chem. 2017, 17, 1777–1785. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Costa, F.; Slivinski, J.; Ióca, L.P.; Bertonha, A.F.; de Felício, R.; Cunha, M.G.D.; da Mata Madeira, P.V.; Cauz, A.C.G.; Trindade, D.M.; Freire, V.F.; et al. Merulinic acid C overcomes gentamicin resistance in Enterococcus faecium. Bioorg. Chem. 2020, 100, 103921. [Google Scholar] [CrossRef]
- Macheboeuf, P.; Fischer, D.S.; Brown, T., Jr.; Zervosen, A.; Luxen, A.; Joris, B.; Dessen, A.; Schofield, C.J. Structural and mechanistic basis of penicillin-binding protein inhibition by lactivicins. Nat. Chem. Biol. 2007, 3, 565–569. [Google Scholar] [CrossRef]








| Bioassay | Source Bacteria | Growth Condition | Bioactive Sample | Bioactivity | |
|---|---|---|---|---|---|
| E. coli | B. subtilis | ||||
| bacterial growth inhibition | LAMA639 | kanamycin | Crude extract | 64 1 (74%) 2 | n.d. 1(<40%) 2 |
| Bacterial growth inhibition | LAMA639 | kanamycin | Fraction 08 | 4 1 (100%) 2 | 2 1 (99%) 2 |
| bacterial growth inhibition | B002_754 | EDTA | Crude extract | n.d.1(<40%) 2 | n.d. 1 (48%) |
| bacterial growth inhibition | B002_754 | EDTA | Fraction 09 | n.d. (<40%) 2 | 8 (99%) 2 |
| ChTL h20S | |||||
| proteasome | LAMA915 | control | Crude extract | n.d. 3 (0%) 4 | |
| proteasome | LAMA915 | control | Fraction 06 | 25 3 (55%) 4 | |
| proteasome | LAMA915 | procaine | Crude extract | n.d. 3 (0%) 4 | |
| proteasome | LAMA915 | procaine | Fraction 06 | 16 3 (0%) 4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Felício, R.; Ballone, P.; Bazzano, C.F.; Alves, L.F.G.; Sigrist, R.; Infante, G.P.; Niero, H.; Rodrigues-Costa, F.; Fernandes, A.Z.N.; Tonon, L.A.C.; et al. Chemical Elicitors Induce Rare Bioactive Secondary Metabolites in Deep-Sea Bacteria under Laboratory Conditions. Metabolites 2021, 11, 107. https://doi.org/10.3390/metabo11020107
de Felício R, Ballone P, Bazzano CF, Alves LFG, Sigrist R, Infante GP, Niero H, Rodrigues-Costa F, Fernandes AZN, Tonon LAC, et al. Chemical Elicitors Induce Rare Bioactive Secondary Metabolites in Deep-Sea Bacteria under Laboratory Conditions. Metabolites. 2021; 11(2):107. https://doi.org/10.3390/metabo11020107
Chicago/Turabian Stylede Felício, Rafael, Patricia Ballone, Cristina Freitas Bazzano, Luiz F. G. Alves, Renata Sigrist, Gina Polo Infante, Henrique Niero, Fernanda Rodrigues-Costa, Arthur Zanetti Nunes Fernandes, Luciane A. C. Tonon, and et al. 2021. "Chemical Elicitors Induce Rare Bioactive Secondary Metabolites in Deep-Sea Bacteria under Laboratory Conditions" Metabolites 11, no. 2: 107. https://doi.org/10.3390/metabo11020107
APA Stylede Felício, R., Ballone, P., Bazzano, C. F., Alves, L. F. G., Sigrist, R., Infante, G. P., Niero, H., Rodrigues-Costa, F., Fernandes, A. Z. N., Tonon, L. A. C., Paradela, L. S., Costa, R. K. E., Dias, S. M. G., Dessen, A., Telles, G. P., da Silva, M. A. C., Lima, A. O. d. S., & Trivella, D. B. B. (2021). Chemical Elicitors Induce Rare Bioactive Secondary Metabolites in Deep-Sea Bacteria under Laboratory Conditions. Metabolites, 11(2), 107. https://doi.org/10.3390/metabo11020107