The Intriguing Role of Iron-Sulfur Clusters in the CIAPIN1 Protein Family
Abstract
:1. Introduction
2. Sequence Analysis of the CIAPIN1 Protein Family
3. Functional Role of the CIAPIN1 Protein Family
3.1. The Maturation of Cytosolic and Nuclear [4Fe-4S] Proteins
3.2. The Diferric-Tyrosyl Radical Cofactor Biosynthesis in Ribonucleotide Reductase
3.3. The Dre2-Tah18 Dependent Regulation of Cell Death
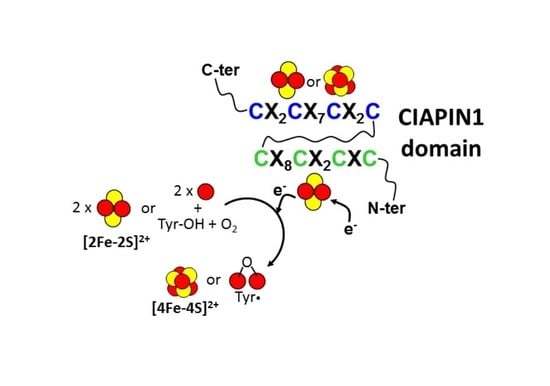
4. Molecular Function of the Iron-Sulfur Cluster Bound to the CX8CX2CXC Motif in the CIAPIN1 Protein Family
4.1. CIAPIN1-Dependent Electron Transfer in the Assembly of [4Fe-4S] Clusters
4.2. CIAPIN1-Dependent Electron Transfer in the Assembly of the Fe(III)2-Y• Cofactor
4.3. A Possible Role of CIAPIN1-Dependent Electron Transfer in the Yeast Viability
4.4. A Possible New Functional Role of the Reducing System Ndor1-Anamorsin
5. Spectroscopic Investigations to Unravel the Nature of the Iron-Sulfur Clusters Bound to CIAPIN1 Proteins
6. Iron-Sulfur Cluster Insertion into CIAPIN1 Proteins
6.1. The Possible Role of Monothiol Glutaredoxins Grx3/4 and Fra2 in Fe/S Cluster Insertion into Dre2
6.2. The Role of Monothiol Glutaredoxin GLRX3 and BOLA2 in [2Fe-2S] Cluster Insertion into Anamorsin
6.3. Cellular Scenario of [2Fe-2S] Cluster Insertion into Anamorsin
7. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beinert, H.; Holm, R.H.; Munck, E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Beinert, H. Iron-sulfur proteins: Ancient structures, still full of surprises. J. Biol. Inorg. Chem. 2000, 5, 2–15. [Google Scholar] [CrossRef]
- Holm, R.H.; Lo, W. Structural Conversions of Synthetic and Protein-Bound Iron-Sulfur Clusters. Chem. Rev. 2016, 116, 13685–13713. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Freibert, S.A.; Rakwalska-Bange, M.; Lill, R. Mechanistic concepts of iron-sulfur protein biogenesis in Biology. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118863. [Google Scholar] [CrossRef] [PubMed]
- Maio, N.; Rouault, T.A. Outlining the Complex Pathway of Mammalian Fe-S Cluster Biogenesis. Trends Biochem. Sci. 2020, 45, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A. Biogenesis of iron-sulfur clusters in mammalian cells: New insights and relevance to human disease. Dis. Model. Mech. 2012, 5, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Sheftel, A.; Stehling, O.; Lill, R. Iron-sulfur proteins in health and disease. Trends Endocrinol. Metab. 2010, 21, 302–314. [Google Scholar] [CrossRef]
- Fontecave, M.; Py, B.; Ollagnier de Choudens, S.; Barras, F. From Iron and Cysteine to Iron-Sulfur Clusters: The Biogenesis Protein Machineries. EcoSal Plus 2008, 3. [Google Scholar] [CrossRef]
- Baussier, C.; Fakroun, S.; Aubert, C.; Dubrac, S.; Mandin, P.; Py, B.; Barras, F. Making iron-sulfur cluster: Structure, regulation and evolution of the bacterial ISC system. Adv. Microb. Physiol. 2020, 76, 1–39. [Google Scholar]
- Garcia, P.S.; Gribaldo, S.; Py, B.; Barras, F. The SUF system: An ABC ATPase-dependent protein complex with a role in Fe-S cluster biogenesis. Res. Microbiol. 2019, 170, 426–434. [Google Scholar] [CrossRef] [Green Version]
- Outten, F.W. Recent advances in the Suf Fe-S cluster biogenesis pathway: Beyond the Proteobacteria. Biochim. Biophys. Acta 2015, 1853, 1464–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc, B.; Gerez, C.; de Choudens, S.O. Assembly of Fe/S proteins in bacterial systems: Biochemistry of the bacterial ISC system. Biochim. Biophys. Acta 2015, 1853, 1436–1447. [Google Scholar] [CrossRef] [PubMed]
- Netz, D.J.; Mascarenhas, J.; Stehling, O.; Pierik, A.J.; Lill, R. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol. 2014, 24, 303–312. [Google Scholar] [CrossRef]
- Zhang, Y.; Lyver, E.R.; Nakamaru-Ogiso, E.; Yoon, H.; Amutha, B.; Lee, D.W.; Bi, E.; Ohnishi, T.; Daldal, F.; Pain, D.; et al. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 2008, 28, 5569–5582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netz, D.J.; Stumpfig, M.; Dore, C.; Muhlenhoff, U.; Pierik, A.J.; Lill, R. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 2010, 6, 758–765. [Google Scholar] [CrossRef]
- Basu, S.; Netz, D.J.; Haindrich, A.C.; Herlerth, N.; Lagny, T.J.; Pierik, A.J.; Lill, R.; Lukeš, J. Cytosolic iron-sulphur protein assembly is functionally conserved and essential in procyclic and bloodstream Trypanosoma brucei. Mol. Microbiol. 2014, 93, 897–910. [Google Scholar] [CrossRef]
- Shibayama, H.; Takai, E.; Matsumura, I.; Kouno, M.; Morii, E.; Kitamura, Y.; Takeda, J.; Kanakura, Y. Identification of a cytokine-induced antiapoptotic molecule anamorsin essential for definitive hematopoiesis. J. Exp. Med. 2004, 199, 581–592. [Google Scholar] [CrossRef] [Green Version]
- Soler, N.; Craescu, C.T.; Gallay, J.; Frapart, Y.M.; Mansuy, D.; Raynal, B.; Baldacci, G.; Pastore, A.; Huang, M.E.; Vernis, L. A S-adenosylmethionine methyltransferase-like domain within the essential, Fe-S-containing yeast protein Dre2. FEBS J. 2012, 279, 2108–2119. [Google Scholar] [CrossRef] [Green Version]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Boscaro, F.; Chatzi, A.; Mikolajczyk, M.; Tokatlidis, K.; Winkelmann, J. Anamorsin is a 2Fe2S cluster-containing substrate of the Mia40-dependent mitochondrial protein trapping machinery. Chem. Biol. 2011, 18, 794–804. [Google Scholar] [CrossRef] [Green Version]
- Banci, L.; Bertini, I.; Calderone, V.; Ciofi-Baffoni, S.; Giachetti, A.; Jaiswal, D.; Mikolajczyk, M.; Piccioli, M.; Winkelmann, J. Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 7136–7141. [Google Scholar] [CrossRef] [Green Version]
- Torruella, G.; Derelle, R.; Paps, J.; Lang, B.F.; Roger, A.J.; Shalchian-Tabrizi, K.; Ruiz-Trillo, I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol. Biol. Evol. 2012, 29, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Bachvaroff, T.R.; Delwiche, C.F. Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 2012, 7, e29696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferecatu, I.; Goncalves, S.; Golinelli-Cohen, M.P.; Clemancey, M.; Martelli, A.; Riquier, S.; Guittet, E.; Latour, J.M.; Puccio, H.; Drapier, J.C.; et al. The diabetes drug target MitoNEET governs a novel trafficking pathway to rebuild an Fe-S cluster into cytosolic aconitase/iron regulatory protein 1. J. Biol. Chem. 2014, 289, 28070–28086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balk, J.; Pierik, A.J.; Netz, D.J.; Muhlenhoff, U.; Lill, R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 2004, 23, 2105–2115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stehling, O.; Jeoung, J.H.; Freibert, S.A.; Paul, V.D.; Bänfer, S.; Niggemeyer, B.; Rösser, R.; Dobbek, H.; Lill, R. Function and crystal structure of the dimeric P-loop ATPase CFD1 coordinating an exposed [4Fe-4S] cluster for transfer to apoproteins. Proc. Natl. Acad. Sci. USA 2018, 115, E9085–E9094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peleh, V.; Riemer, J.; Dancis, A.; Herrmann, J.M. Protein oxidation in the intermembrane space of mitochondria is substrate-specific rather than general. Microb. Cell 2014, 1, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Vernis, L.; Facca, C.; Delagoutte, E.; Soler, N.; Chanet, R.; Guiard, B.; Faye, G.; Baldacci, G. A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS ONE 2009, 4, e4376. [Google Scholar] [CrossRef] [Green Version]
- Soler, N.; Delagoutte, E.; Miron, S.; Facca, C.; Baille, D.; d’Autreaux, B.; Craescu, G.; Frapart, Y.M.; Mansuy, D.; Baldacci, G.; et al. Interaction between the reductase Tah18 and highly conserved Fe-S containing Dre2 C-terminus is essential for yeast viability. Mol. Microbiol. 2011, 82, 54–67. [Google Scholar] [CrossRef]
- Frey, A.G.; Palenchar, D.J.; Wildemann, J.D.; Philpott, C.C. A Glutaredoxin-BolA Complex Serves as an Iron-Sulfur Cluster Chaperone for the Cytosolic Cluster Assembly Machinery. J. Biol. Chem. 2016, 291, 22344–22356. [Google Scholar] [CrossRef] [Green Version]
- Stubbe, J.; van Der Donk, W.A. Protein Radicals in Enzyme Catalysis. Chem. Rev. 1998, 98, 705–762. [Google Scholar] [CrossRef]
- Nordlund, P.; Reichard, P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006, 75, 681–706. [Google Scholar] [CrossRef] [PubMed]
- Chabes, A.; Domkin, V.; Larsson, G.; Liu, A.; Graslund, A.; Wijmenga, S.; Thelander, L. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc. Natl. Acad. Sci. USA 2000, 97, 2474–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, L.; Wu, X.; An, X.; Stubbe, J.; Huang, M. Investigation of in vivo diferric tyrosyl radical formation in Saccharomyces cerevisiae Rnr2 protein: Requirement of Rnr4 and contribution of Grx3/4 AND Dre2 proteins. J. Biol. Chem. 2011, 286, 41499–41509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, H.; Zhang, C.; An, X.; Liu, L.; Stubbe, J.; Huang, M. Conserved electron donor complex Dre2-Tah18 is required for ribonucleotide reductase metallocofactor assembly and DNA synthesis. Proc. Natl. Acad. Sci. USA 2014, 111, E1695–E1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Stumpfig, M.; Zhang, C.; An, X.; Stubbe, J.; Lill, R.; Huang, M. The diferric-tyrosyl radical cluster of ribonucleotide reductase and cytosolic iron-sulfur clusters have distinct and similar biogenesis requirements. J. Biol. Chem. 2017, 292, 11445–11451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshikawa, Y.; Nasuno, R.; Kawahara, N.; Nishimura, A.; Watanabe, D.; Takagi, H. Regulatory mechanism of the flavoprotein Tah18-dependent nitric oxide synthesis and cell death in yeast. Nitric Oxide 2016, 57, 85–91. [Google Scholar] [CrossRef]
- Almeida, B.; Buttner, S.; Ohlmeier, S.; Silva, A.; Mesquita, A.; Sampaio-Marques, B.; Osório, N.S.; Kollau, A.; Mayer, B.; Leão, C.; et al. NO-mediated apoptosis in yeast. J. Cell Sci. 2007, 120, 3279–3288. [Google Scholar] [CrossRef] [Green Version]
- Murataliev, M.B.; Feyereisen, R.; Walker, F.A. Electron transfer by diflavin reductases. Biochim. Biophys. Acta 2004, 1698, 1–26. [Google Scholar] [CrossRef]
- Banci, L.; Ciofi-Baffoni, S.; Mikolajczyk, M.; Winkelmann, J.; Bill, E.; Pandelia, M.E. Human anamorsin binds [2Fe-2S] clusters with unique electronic properties. J. Biol. Inorg. Chem. 2013, 18, 883–893. [Google Scholar] [CrossRef]
- Paul, V.D.; Lill, R. Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 2015, 1853, 1528–1539. [Google Scholar] [CrossRef] [Green Version]
- Kunichika, K.; Nakamura, R.; Fujishiro, T.; Takahashi, Y. The Structure of the Dimeric State of IscU Harboring Two Adjacent [2Fe-2S] Clusters Provides Mechanistic Insights into Cluster Conversion to [4Fe-4S]. Biochemistry 2021, 60, 1569–1572. [Google Scholar] [CrossRef] [PubMed]
- Azam, T.; Przybyla-Toscano, J.; Vignols, F.; Couturier, J.; Rouhier, N.; Johnson, M.K. The Arabidopsis Mitochondrial Glutaredoxin GRXS15 Provides [2Fe-2S] Clusters for ISCA-Mediated [4Fe-4S] Cluster Maturation. Int. J. Mol. Sci. 2020, 21, 9237. [Google Scholar] [CrossRef] [PubMed]
- Weiler, B.D.; Brück, M.C.; Kothe, I.; Bill, E.; Lill, R.; Mühlenhoff, U. Mitochondrial [4Fe-4S] protein assembly involves reductive [2Fe-2S] cluster fusion on ISCA1-ISCA2 by electron flow from ferredoxin FDX2. Proc. Natl. Acad. Sci. USA 2020, 117, 20555–20565. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, K.; Unciuleac, M.C.; Naik, S.; Dean, D.R.; Huynh, B.H.; Johnson, M.K. Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry 2007, 46, 6804–6811. [Google Scholar] [CrossRef]
- Brancaccio, D.; Gallo, A.; Mikolajczyk, M.; Zovo, K.; Palumaa, P.; Novellino, E.; Piccioli, M.; Ciofi-Baffoni, S.; Banci, L. Formation of [4Fe-4S] clusters in the mitochondrial iron-sulfur cluster assembly machinery. J. Am. Chem. Soc. 2014, 136, 16240–16250. [Google Scholar] [CrossRef]
- Brancaccio, D.; Gallo, A.; Piccioli, M.; Novellino, E.; Ciofi-Baffoni, S.; Banci, L. [4Fe-4S] Cluster Assembly in Mitochondria and Its Impairment by Copper. J. Am. Chem. Soc. 2017, 139, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Laursen, T.; Jensen, K.; Moller, B.L. Conformational changes of the NADPH-dependent cytochrome P450 reductase in the course of electron transfer to cytochromes P450. Biochim. Biophys. Acta 2011, 1814, 132–138. [Google Scholar] [CrossRef]
- Ellis, J.; Gutierrez, A.; Barsukov, I.L.; Huang, W.C.; Grossmann, J.G.; Roberts, G.C. Domain motion in cytochrome P450 reductase: Conformational equilibria revealed by NMR and small-angle x-ray scattering. J. Biol. Chem. 2009, 284, 36628–36637. [Google Scholar] [CrossRef] [Green Version]
- Tarassov, K.; Messier, V.; Landry, C.R.; Radinovic, S.; Serna Molina, M.M.; Shames, I.; Malitskaya, Y.; Vogel, J.; Bussey, H.; Michnick, S.W. An in vivo map of the yeast protein interactome. Science 2008, 320, 1465–1470. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Mapolelo, D.T.; Dingra, N.N.; Naik, S.G.; Lees, N.S.; Hoffman, B.M.; Riggs-Gelasco, P.J.; Huynh, B.H.; Johnson, M.K.; Outten, C.E. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 2009, 48, 9569–9581. [Google Scholar] [CrossRef] [Green Version]
- Muhlenhoff, U.; Molik, S.; Godoy, J.R.; Uzarska, M.A.; Richter, N.; Seubert, A.; Zhang, Y.; Stubbe, J.; Pierrel, F.; Herrero, E.; et al. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010, 12, 373–385. [Google Scholar] [CrossRef] [Green Version]
- Camponeschi, F.; Ciofi-Baffoni, S.; Banci, L. Anamorsin/Ndor1 Complex Reduces [2Fe-2S]-MitoNEET via a Transient Protein-Protein Interaction. J. Am. Chem. Soc. 2017, 139, 9479–9482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamir, S.; Paddock, M.L.; Darash-Yahana-Baram, M.; Holt, S.H.; Sohn, Y.S.; Agranat, L.; Michaeli, D.; Stofleth, J.T.; Lipper, C.H.; Morcos, F.; et al. Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim. Biophys. Acta 2015, 1853, 1294–1315. [Google Scholar] [CrossRef] [Green Version]
- Karmi, O.; Marjault, H.B.; Pesce, L.; Carloni, P.; Onuchic, J.N.; Jennings, P.A.; Mittler, R.; Nechushtai, R. The unique fold and lability of the [2Fe-2S] clusters of NEET proteins mediate their key functions in health and disease. J. Biol. Inorg. Chem. 2018, 23, 599–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bak, D.W.; Elliott, S.J. Conserved hydrogen bonding networks of MitoNEET tune Fe-S cluster binding and structural stability. Biochemistry 2013, 52, 4687–4696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuris, J.A.; Harir, Y.; Conlan, A.R.; Shvartsman, M.; Michaeli, D.; Tamir, S.; Paddock, M.L.; Onuchic, J.N.; Mittler, R.; Cabantchik, Z.I.; et al. Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc. Natl. Acad. Sci. USA 2011, 108, 13047–13052. [Google Scholar] [CrossRef] [Green Version]
- Golinelli-Cohen, M.P.; Lescop, E.; Mons, C.; Goncalves, S.; Clemancey, M.; Santolini, J.; Guittet, E.; Blondin, G.; Latour, J.M.; Bouton, C. Redox Control of the Human Iron-Sulfur Repair Protein MitoNEET Activity via Its Iron-Sulfur Cluster. J. Biol. Chem. 2016, 291, 7583–7593. [Google Scholar] [CrossRef] [Green Version]
- Landry, A.P.; Ding, H. Redox control of human mitochondrial outer membrane protein MitoNEET [2Fe-2S] clusters by biological thiols and hydrogen peroxide. J. Biol. Chem. 2014, 289, 4307–4315. [Google Scholar] [CrossRef] [Green Version]
- Bak, D.W.; Zuris, J.A.; Paddock, M.L.; Jennings, P.A.; Elliott, S.J. Redox characterization of the FeS protein MitoNEET and impact of thiazolidinedione drug binding. Biochemistry 2009, 48, 10193–10195. [Google Scholar] [CrossRef] [Green Version]
- Conlan, A.R.; Axelrod, H.L.; Cohen, A.E.; Abresch, E.C.; Zuris, J.; Yee, D.; Nechushtai, R.; Jennings, P.A.; Paddock, M.L. Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram Syndrome 2. J. Mol. Biol. 2009, 392, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Stehling, O.; Mascarenhas, J.; Vashisht, A.A.; Sheftel, A.D.; Niggemeyer, B.; Rosser, R.; Pierik, A.J.; Wohlschlegel, J.A.; Lill, R. Human CIA2A-FAM96A and CIA2B-FAM96B integrate iron homeostasis and maturation of different subsets of cytosolic-nuclear iron-sulfur proteins. Cell Metab. 2013, 18, 187–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, G.; Liu, D.; Pan, F.; Zhao, J.; Li, T.; Ma, Y.; Shen, B.; Lyu, J. His-87 ligand in mitoNEET is crucial for the transfer of iron sulfur clusters from mitochondria to cytosolic aconitase. Biochem. Biophy.s Res. Commun. 2016, 470, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Mons, C.; Ferecatu, I.; Riquier, S.; Lescop, E.; Bouton, C.; Golinelli-Cohen, M.P. Combined Biochemical, Biophysical, and Cellular Methods to Study Fe-S Cluster Transfer and Cytosolic Aconitase Repair by MitoNEET. Methods Enzym. 2017, 595, 83–106. [Google Scholar]
- Kühn, L.C. Iron regulatory proteins and their role in controlling iron metabolism. Metallomics 2015, 7, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, C.; Dancis, A.; Nakamaru-Ogiso, E. EPR studies of wild type and mutant Dre2 identify essential [2Fe--2S] and [4Fe--4S] clusters and their cysteine ligands. J. Biochem. 2017, 161, 67–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netz, D.J.; Genau, H.M.; Weiler, B.D.; Bill, E.; Pierik, A.J.; Lill, R. The conserved protein Dre2 uses essential [2Fe-2S] and [4Fe-4S] clusters for its function in cytosolic iron-sulfur protein assembly. Biochem. J. 2016, 473, 2073–2085. [Google Scholar] [CrossRef]
- Bernard, D.G.; Netz, D.J.; Lagny, T.J.; Pierik, A.J.; Balk, J. Requirements of the cytosolic iron-sulfur cluster assembly pathway in Arabidopsis. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matteucci, S.; Camponeschi, F.; Clémancey, M.; Ciofi-Baffoni, S.; Blondin, G.; Banci, L. In Cellulo Mössbauer and EPR Studies Bring New Evidence to the Long-Standing Debate on Iron-Sulfur Cluster Binding in Human Anamorsin. Angew. Chem. Int. Ed. 2021, 60, 14841–14845. [Google Scholar] [CrossRef] [PubMed]
- Hagen, W.R. EPR spectroscopy of complex biological iron-sulfur systems. J. Biol. Inorg. Chem. 2018, 23, 623–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blondin, G.; Girerd, J.-J. Interplay of electron exchange and electron transfer in metal polynuclear complexes in proteins or chemical models. Chem. Rev. 1990, 90, 1359–1376. [Google Scholar] [CrossRef]
- Iwema, T.; Picciocchi, A.; Traore, D.A.; Ferrer, J.L.; Chauvat, F.; Jacquamet, L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry 2009, 48, 6041–6043. [Google Scholar] [CrossRef] [PubMed]
- Picciocchi, A.; Saguez, C.; Boussac, A.; Cassier-Chauvat, C.; Chauvat, F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry 2007, 46, 15018–15026. [Google Scholar] [CrossRef] [PubMed]
- Kumanovics, A.; Chen, O.S.; Li, L.; Bagley, D.; Adkins, E.M.; Lin, H.; Dingra, N.N.; Outten, C.E.; Keller, G.; Winge, D.; et al. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 2008, 283, 10276–10286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talib, E.A.; Outten, C.E. Iron-sulfur cluster biogenesis, trafficking, and signaling: Roles for CGFS glutaredoxins and BolA proteins. Biochim. Biophys. Acta. Mol. Cell Res. 2020, 1868, 118847. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Outten, C.E. Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry 2012, 51, 4377–4389. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Mapolelo, D.T.; Dingra, N.N.; Keller, G.; Riggs-Gelasco, P.J.; Winge, D.R.; Johnson, M.K.; Outten, C.E. Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast. J. Biol. Chem. 2011, 286, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Chi, C.B.; Tang, Y.; Zhang, J.; Dai, Y.N.; Abdalla, M.; Chen, Y.; Zhou, C.Z. Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3. J. Mol. Biol. 2018, 430, 1235–1248. [Google Scholar] [CrossRef]
- Pandey, A.K.; Pain, J.; Dancis, A.; Pain, D. Mitochondria export iron-sulfur and sulfur intermediates to the cytoplasm for iron-sulfur cluster assembly and tRNA thiolation in yeast. J. Biol. Chem. 2019, 294, 9489–9502. [Google Scholar] [CrossRef]
- Freibert, S.A.; Boniecki, M.T.; Stümpfig, C.; Schulz, V.; Krapoth, N.; Winge, D.R.; Mühlenhoff, U.; Stehling, O.; Cygler, M.; Lill, R. N-terminal tyrosine of ISCU2 triggers [2Fe-2S] cluster synthesis by ISCU2 dimerization. Nat. Commun. 2021, 12, 6902. [Google Scholar] [CrossRef]
- Poor, C.B.; Wegner, S.V.; Li, H.; Dlouhy, A.C.; Schuermann, J.P.; Sanishvili, R.; Hinshaw, J.R.; Riggs-Gelasco, P.J.; Outten, C.E.; He, C. Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc. Natl. Acad. Sci. USA 2014, 111, 4043–4048. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.; Shibayama, H.; Tanaka, H.; Tanimura, A.; Matsumura, I.; Kanakura, Y. PICOT is a molecule which binds to anamorsin. Biochem. Biophys. Res. Commun. 2011, 408, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Haunhorst, P.; Berndt, C.; Eitner, S.; Godoy, J.R.; Lillig, C.H. Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein. Biochem. Biophys. Res. Commun. 2010, 394, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Ciofi-Baffoni, S.; Gajda, K.; Muzzioli, R.; Peruzzini, R.; Winkelmann, J. N-terminal domains mediate [2Fe-2S] cluster transfer from glutaredoxin-3 to anamorsin. Nat. Chem. Biol. 2015, 11, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Haunhorst, P.; Hanschmann, E.M.; Brautigam, L.; Stehling, O.; Hoffmann, B.; Muhlenhoff, U.; Lill, R.; Berndt, C.; Lillig, C.H. Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol. Biol. Cell 2013, 24, 1895–1903. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Li, H.; Mapolelo, D.T.; Randeniya, S.; Johnson, M.K.; Outten, C.E. Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry 2012, 51, 1687–1696. [Google Scholar] [CrossRef]
- Banci, L.; Camponeschi, F.; Ciofi-Baffoni, S.; Muzzioli, R. Elucidating the molecular function of human BOLA2 in GRX3-Dependent anamorsin maturation pathway. J. Am. Chem. Soc. 2015, 137, 16133–16134. [Google Scholar] [CrossRef]
- Gibson, L.M.; Dingra, N.N.; Outten, C.E.; Lebioda, L. Structure of the thioredoxin-like domain of yeast glutaredoxin 3. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 927–932. [Google Scholar] [CrossRef]
- Martin, J.L.; McMillan, F.M. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002, 12, 783–793. [Google Scholar] [CrossRef]
- Philpott, C.C.; Ryu, M.S.; Frey, A.; Patel, S. Cytosolic iron chaperones: Proteins delivering iron cofactors in the cytosol of mammalian cells. J. Biol. Chem. 2017, 292, 12764–12771. [Google Scholar] [CrossRef] [Green Version]
- Maio, N.; Rouault, T.A. Mammalian iron sulfur cluster biogenesis: From assembly to delivery to recipient proteins with a focus on novel targets of the chaperone and co-chaperone proteins. IUBMB Life 2022. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Bencze, K.Z.; Stemmler, T.L.; Philpott, C.C. A cytosolic iron chaperone that delivers iron to ferritin. Science 2008, 320, 1207–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makeyev, A.V.; Liebhaber, S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. Rna 2002, 8, 265–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.J.; Frey, A.G.; Palenchar, D.J.; Achar, S.; Bullough, K.Z.; Vashisht, A.; Wohlschlegel, J.A.; Philpott, C.C. A PCBP1-BolA2 chaperone complex delivers iron for cytosolic [2Fe-2S] cluster assembly. Nat. Chem. Biol. 2019, 15, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.J.; Protchenko, O.; Shakoury-Elizeh, M.; Baratz, E.; Jadhav, S.; Philpott, C.C. The iron chaperone and nucleic acid-binding activities of poly(rC)-binding protein 1 are separable and independently essential. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Py, B.; Gerez, C.; Angelini, S.; Planel, R.; Vinella, D.; Loiseau, L.; Talla, E.; Brochier-Armanet, C.; Garcia, S.R.; Latour, J.M.; et al. Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol. Microbiol. 2012, 86, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Unciuleac, M.C.; Dean, D.R. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J. Bacteriol. 2006, 188, 7551–7561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, G.; Lu, J.; Bitoun, J.P.; Huang, H.; Ding, H. IscA/SufA paralogues are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem. J. 2009, 420, 463–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelini, S.; Gerez, C.; Ollagnier-de, C.S.; Sanakis, Y.; Fontecave, M.; Barras, F.; Py, B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 2008, 283, 14084–14091. [Google Scholar] [CrossRef] [Green Version]
- Bandyopadhyay, S.; Naik, S.G.; O’Carroll, I.P.; Huynh, B.H.; Dean, D.R.; Johnson, M.K.; Dos Santos, P.C. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J. Biol. Chem. 2008, 283, 14092–14099. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.M.; Freire, P.; Vicente, M.; Arraiano, C.M. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol. Microbiol. 1999, 32, 789–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huynen, M.A.; Spronk, C.A.; Gabaldón, T.; Snel, B. Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS Lett. 2005, 579, 591–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreini, C.; Rosato, A.; Banci, L. The relationship between environmental dioxygen and iron-sulfur proteins explored at the genome level. PLoS ONE 2017, 12, e0171279. [Google Scholar] [CrossRef] [PubMed]
- Willems, P.; Wanschers, B.F.; Esseling, J.; Szklarczyk, R.; Kudla, U.; Duarte, I.; Forkink, M.; Nooteboom, M.; Swarts, H.; Gloerich, J.; et al. BOLA1 is an aerobic protein that prevents mitochondrial morphology changes induced by glutathione depletion. Antioxid. Redox. Signal. 2013, 18, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Paul, V.D.; Muhlenhoff, U.; Stumpfig, M.; Seebacher, J.; Kugler, K.G.; Renicke, C.; Taxis, C.; Gavin, A.C.; Pierik, A.J.; Lill, R. The deca-GX3 proteins Yae1-Lto1 function as adaptors recruiting the ABC protein Rli1 for iron-sulfur cluster insertion. Elife 2015, 4, e08231. [Google Scholar] [CrossRef]
- Nuttle, X.; Giannuzzi, G.; Duyzend, M.H.; Schraiber, J.G.; Narvaiza, I.; Camponeschi, F.; Ciofi-Baffoni, S.; Sudmant, P.H.; Penn, O.; Chiatante, G.; et al. Emergence of a Homo sapiens-specific gene family and the evolution of autism risk at chromosome 16p11.2. Nature 2016, 536, 205–209. [Google Scholar] [CrossRef] [Green Version]
- Lipper, C.H.; Paddock, M.L.; Onuchic, J.N.; Mittler, R.; Nechushtai, R.; Jennings, P.A. Cancer-Related NEET Proteins Transfer 2Fe-2S Clusters to Anamorsin, a Protein Required for Cytosolic Iron-Sulfur Cluster Biogenesis. PLoS ONE 2015, 10, e0139699. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Clade | # Organisms per Clade | # Organisms with One CIAPIN1 Protein | # Organisms with More Than One CIAPIN1 Protein | Domain Architecture |
|---|---|---|---|---|
| Amorphea | 1214 | 1080 | 134 | Ciapin1_only + Methyltransf11_Ciapin1 + Dre2_Ciapin1 |
| Cryptista | 1 | 0 | 1 | Ciapin1_only |
| Discoba | 18 | 17 | 1 | Ciapin1_only * |
| Haptista | 1 | 1 | 0 | Ciapin1_only |
| TSar | 91 | 82 | 9 | Ciapin1_only |
| Archaeplastida | 185 | 83 | 102 | Ciapin1_only + Methyltransf11_Ciapin1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciofi-Baffoni, S.; Andreini, C. The Intriguing Role of Iron-Sulfur Clusters in the CIAPIN1 Protein Family. Inorganics 2022, 10, 52. https://doi.org/10.3390/inorganics10040052
Ciofi-Baffoni S, Andreini C. The Intriguing Role of Iron-Sulfur Clusters in the CIAPIN1 Protein Family. Inorganics. 2022; 10(4):52. https://doi.org/10.3390/inorganics10040052
Chicago/Turabian StyleCiofi-Baffoni, Simone, and Claudia Andreini. 2022. "The Intriguing Role of Iron-Sulfur Clusters in the CIAPIN1 Protein Family" Inorganics 10, no. 4: 52. https://doi.org/10.3390/inorganics10040052
APA StyleCiofi-Baffoni, S., & Andreini, C. (2022). The Intriguing Role of Iron-Sulfur Clusters in the CIAPIN1 Protein Family. Inorganics, 10(4), 52. https://doi.org/10.3390/inorganics10040052