Synthesis of an Aryl-Semicarbazone-Based Cu(II) Complex for DNA and BSA Interaction and Anti-Cancer Activity against Human Cervix Uteri Carcinoma
Abstract
:1. Introduction
2. Results and Discussions
2.1. Synthesis
2.2. Spectroscopic Studies
2.2.1. UV−Vis Spectroscopy
2.2.2. FTIR Spectroscopy
2.2.3. Mass Spectroscopy
2.3. Theoretical Analysis
2.4. Solution Stability and Lipophilicity
2.5. DNA Binding
2.5.1. Electronic Absorption
2.5.2. Emission Studies
2.5.3. Determination of Enthalpy and Entropy Changes (ΔH and ΔS)
2.5.4. Binding Mode
2.6. Binding Affinity with BSA
Enthalpy (ΔH) and Entropy (ΔS) Determination
2.7. Molecular Docking with DNA and BSA
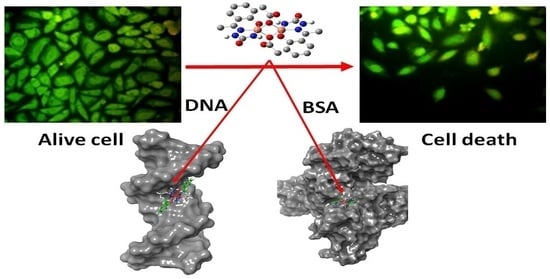
2.8. Cytotoxicity Studies with SiHa Cell Lines
3. Experimental Section
3.1. Materials
3.2. Ligand Preparation
3.3. Copper(II) Complex
3.4. Physical Measurements
3.5. Theoretical Studies
3.6. Stability Evaluation
3.7. Lipophilicity Check
3.8. DNA Binding
3.9. Interaction with BSA
3.10. Cytotoxicity
3.10.1. Culture and Maintenance of SiHa Cells
3.10.2. Viability of the Cells
3.10.3. Fluorescence Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.P.M. Platinum and Palladium Polyamine Complexes as Anticancer Agents: The Structural Factor. ISRN Spectrosc. 2013, 2013, 287353. [Google Scholar] [CrossRef]
- Abu-Surrah, A.; Kettunen, M. Platinum Group Antitumor Chemistry: Design and development of New Anticancer Drugs Complementary to Cisplatin. Curr. Med. Chem. 2006, 13, 1337–1357. [Google Scholar] [CrossRef] [PubMed]
- Karami, K.; Mehri Lighvan, Z.; Alizadeh, A.M.; Poshteh-Shirani, M.; Khayamian, T.; Lipkowski, J. Synthesis of a novel trinuclear palladium complex: The influence of an oxime chelate ligand on biological evaluation towards double-strand DNA, BSA protein and molecular modeling studies. RSC Adv. 2016, 6, 78424–78435. [Google Scholar] [CrossRef]
- Raman, N.; Dhaveethu Raja, J.; Sakthivel, A. Synthesis, spectral characterization of Schiff base transition metal complexes: DNA cleavage and antimicrobial activity studies. J. Chem. Sci. 2007, 119, 303–310. [Google Scholar] [CrossRef]
- Ott, I.; Gust, R. Non Platinum Metal Complexes as Anti-cancer Drugs. Arch. Pharm. 2007, 340, 117–126. [Google Scholar] [CrossRef]
- Belicchi Ferrari, M.; Bisceglie, F.; Gasparri Fava, G.; Pelosi, G.; Tarasconi, P.; Albertini, R.; Pinelli, S. Synthesis, characterization and biological activity of two new polymeric copper(II) complexes with α-ketoglutaric acid thiosemicarbazone. J. Inorg. Biochem. 2002, 89, 36–44. [Google Scholar] [CrossRef]
- Ranford, J.D.; Sadler, P.J.; Tocher, D.A. Cytotoxicity and antiviral activity of transition-metal salicylato complexes and crystal structure of Bis(diisopropylsalicylato)(1,10-phenanthroline)copper(II). J. Chem. Soc. Dalt. Trans. 1993, 22, 3393. [Google Scholar] [CrossRef]
- Nonkuntod, P.; Boonmak, J.; Senawong, T.; Soikum, C.; Chaveerach, P.; Watwiangkham, A.; Suthirakun, S.; Chaveerach, U. The effect of gallic acid on the copper(ii) complex of N -(methylpyridin-2-yl)-amidino- O -methylurea: Crystal structure, DNA interactions, in vitro cytotoxicity and antibacterial activity. New J. Chem. 2023, 47, 12259–12273. [Google Scholar] [CrossRef]
- Vilar, S.; Costanzi, S. Predicting the Biological Activities Through QSAR Analysis and Docking-Based Scoring. In Membrane Protein Structure and Dynamics: Methods and Protocols; Springer Nature: Berlin/Heidelberg, Germany, 2012; pp. 271–284. [Google Scholar]
- Mir, I.A.; Ain, Q.U.; Qadir, T.; Malik, A.Q.; Jan, S.; Shahverdi, S.; Nabi, S.A. A review of semicarbazone-derived metal complexes for application in biomedicine and related fields. J. Mol. Struct. 2024, 1295, 136216. [Google Scholar] [CrossRef]
- Liu, R.; Cui, J.; Ding, T.; Liu, Y.; Liang, H. Research Progress on the Biological Activities of Metal Complexes Bearing Polycyclic Aromatic Hydrazones. Molecules 2022, 27, 8393. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.F.; Yang, C.-T.; Fan, D.; Vittal, J.J.; Ranford, J.D. Synthesis, characterization and physicochemical properties of copper(II) complexes containing salicylaldehyde semicarbazone. Polyhedron 2003, 22, 2781–2786. [Google Scholar] [CrossRef]
- Chen, R.; Liu, C.-S.; Zhang, H.; Guo, Y.; Bu, X.-H.; Yang, M. Three new Cu(II) and Cd(II) complexes with 3-(2-pyridyl)pyrazole-based ligand: Syntheses, crystal structures, and evaluations for bioactivities. J. Inorg. Biochem. 2007, 101, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, S.; Al-Asbahy, W.M.; Afzal, M.; Arjmand, F.; Hasan Khan, R. Interaction and photo-induced cleavage studies of a copper based chemotherapeutic drug with human serum albumin: Spectroscopic and molecular docking study. Mol. Biosyst. 2012, 8, 2424. [Google Scholar] [CrossRef]
- Maity, R.; Sepay, N.; Pramanik, U.; Jana, K.; Mukherjee, S.; Maity, S.; Mal, D.; Maity, T.; Samanta, B.C. Exploring the Noncovalent Interactions of the Dinuclear Cu(II) Schiff Base Complex with Bovine Serum Albumin and Cell Viability against the SiHa Cancer Cell Line. J. Phys. Chem. B 2021, 125, 11364–11373. [Google Scholar] [CrossRef]
- Hansch, C.; Björkroth, J.P.; Leo, A. Hydrophobicity and Central Nervous System Agents: On the Principle of Minimal Hydrophobicity in Drug Design. J. Pharm. Sci. 1987, 76, 663–687. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Tian, N.; Zhou, Z.-Y.; Sun, S.-G.; Ding, Y.; Wang, Z.L. Synthesis of Tetrahexahedral Platinum Nanocrystals with High-Index Facets and High Electro-Oxidation Activity. Science 2007, 316, 732–735. [Google Scholar] [CrossRef]
- Cox, P.J.; Psomas, G.; Bolos, C.A. Characterization and DNA-interaction studies of 1,1-dicyano-2,2-ethylene dithiolate Ni(II) mixed-ligand complexes with 2-amino-5-methyl thiazole, 2-amino-2-thiazoline and imidazole. Crystal structure of [Ni(i-MNT)(2a-5mt)2]. Bioorg. Med. Chem. 2009, 17, 6054–6062. [Google Scholar] [CrossRef]
- Jana, K.; Maity, R.; Puschmann, H.; Mitra, A.; Ghosh, R.; Debnath, S.C.; Shukla, A.; Mahanta, A.K.; Maity, T.; Samanta, B.C. A binuclear chloride bridged Cu(II) and a mononuclear Ni(II) complex: Synthesis, crystal structure, photo catalytic and biological studies. Inorganica Chim. Acta 2021, 515, 120067. [Google Scholar] [CrossRef]
- Shahabadi, N.; Kashanian, S.; Darabi, F. In Vitro Study of DNA Interaction with a Water-Soluble Dinitrogen Schiff Base. DNA Cell Biol. 2009, 28, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, L.H.; Abu-Dief, A.M.; El-Khatib, R.M.; Abdel-Fatah, S.M. Sonochemical synthesis, DNA binding, antimicrobial evaluation and in vitro anticancer activity of three new nano-sized Cu(II), Co(II) and Ni(II) chelates based on tri-dentate NOO imine ligands as precursors for metal oxides. J. Photochem. Photobiol. B Biol. 2016, 162, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Lakshmipraba, J.; Arunachalam, S.; Riyasdeen, A.; Dhivya, R.; Akbarsha, M.A. Polyethyleneimine anchored copper(II) complexes: Synthesis, characterization, in vitro DNA binding studies and cytotoxicity studies. J. Photochem. Photobiol. B Biol. 2015, 142, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Ünver, H.; Boyacıoğlu, B.; Zeyrek, C.T.; Yıldız, M.; Demir, N.; Yıldırım, N.; Karaosmanoğlu, O.; Sivas, H.; Elmalı, A. Synthesis, spectral and quantum chemical studies and use of (E)-3-[(3,5-bis(trifluoromethyl)phenylimino)methyl]benzene-1,2-diol and its Ni(II) and Cu(II) complexes as an anion sensor, DNA binding, DNA cleavage, anti-microbial, anti-mutagenic and anti-ca. J. Mol. Struct. 2016, 1125, 162–176. [Google Scholar] [CrossRef]
- Rajendiran, V.; Karthik, R.; Palaniandavar, M.; Stoeckli-Evans, H.; Periasamy, V.S.; Akbarsha, M.A.; Srinag, B.S.; Krishnamurthy, H. Mixed-Ligand Copper(II)-phenolate Complexes: Effect of Coligand on Enhanced DNA and Protein Binding, DNA Cleavage, and Anticancer Activity. Inorg. Chem. 2007, 46, 8208–8221. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Das, S.; Kumar, G.S. Molecular aspects on the interaction of phenosafranine to deoxyribonucleic acid: Model for intercalative drug–DNA binding. J. Mol. Struct. 2008, 872, 56–63. [Google Scholar] [CrossRef]
- Kumar, C.V.; Turner, R.S.; Asuncion, E.H. Groove binding of a styrylcyanine dye to the DNA double helix: The salt effect. J. Photochem. Photobiol. A Chem. 1993, 74, 231–238. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; Alzuet, G.; González-Álvarez, M.; Liu-González, M.; Castiñeiras, A.; Borrás, J. Oxidative nuclease activity of ferromagnetically coupled μ-hydroxo-μ-propionato copper(II) complexes [Cu3(L)2(μ-OH)2(μ-propionato)2] (L=N-(pyrid-2-ylmethyl)R-sulfonamidato, R=benzene, toluene, naphthalene). J. Inorg. Biochem. 2009, 103, 243–255. [Google Scholar] [CrossRef]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug–DNA interactions and their study by UV–Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B Biol. 2013, 124, 1–19. [Google Scholar] [CrossRef]
- Record, M.T.; Anderson, C.F.; Lohman, T.M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: The roles of ion association or release, screening, and ion effects on water activity. Q. Rev. Biophys. 1978, 11, 103–178. [Google Scholar] [CrossRef] [PubMed]
- Lian, W.-J.; Wang, X.-T.; Xie, C.-Z.; Tian, H.; Song, X.-Q.; Pan, H.-T.; Qiao, X.; Xu, J.-Y. Mixed-ligand copper(II) Schiff base complexes: The role of the co-ligand in DNA binding, DNA cleavage, protein binding and cytotoxicity. Dalt. Trans. 2016, 45, 9073–9087. [Google Scholar] [CrossRef] [PubMed]
- Sepay, N.; Banerjee, M.; Islam, R.; Dey, S.P.; Halder, U.C. Crystallography based exploration of non-covalent interactions to design, synthesis of coumarin for stronger protein binding. Phys. Chem. Chem. Phys. 2022, 11, 6605–6615. [Google Scholar] [CrossRef] [PubMed]
- Margiotta, N.; Natile, G.; Capitelli, F.; Fanizzi, F.P.; Boccarelli, A.; De Rinaldis, P.; Giordano, D.; Coluccia, M. Sterically hindered complexes of platinum(II) with planar heterocyclic nitrogen donors. A novel complex with 1-methyl-cytosine has a spectrum of activity different from cisplatin and is able of overcoming acquired cisplatin resistance. J. Inorg. Biochem. 2006, 100, 1849–1857. [Google Scholar] [CrossRef]
- Jagadeesan, S.; Balasubramanian, V.; Baumann, P.; Neuburger, M.; Häussinger, D.; Palivan, C.G. Water-Soluble Co(III) Complexes of Substituted Phenanthrolines with Cell Selective Anticancer Activity. Inorg. Chem. 2013, 52, 12535–12544. [Google Scholar] [CrossRef]








| Temperature (K) | Kb | ΔG (kJ mol−1) |
|---|---|---|
| 298 | 7.128 × 104 | −27.68 |
| 303 | 2.44 × 105 | −31.24 |
| 308 | 4.72 × 105 | −33.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maity, R.; Manna, B.; Maity, S.; Jana, K.; Maity, T.; Afzal, M.; Sepay, N.; Samanta, B.C. Synthesis of an Aryl-Semicarbazone-Based Cu(II) Complex for DNA and BSA Interaction and Anti-Cancer Activity against Human Cervix Uteri Carcinoma. Inorganics 2024, 12, 19. https://doi.org/10.3390/inorganics12010019
Maity R, Manna B, Maity S, Jana K, Maity T, Afzal M, Sepay N, Samanta BC. Synthesis of an Aryl-Semicarbazone-Based Cu(II) Complex for DNA and BSA Interaction and Anti-Cancer Activity against Human Cervix Uteri Carcinoma. Inorganics. 2024; 12(1):19. https://doi.org/10.3390/inorganics12010019
Chicago/Turabian StyleMaity, Ribhu, Biplab Manna, Swapan Maity, Kalyanmoy Jana, Tithi Maity, Mohd Afzal, Nayim Sepay, and Bidhan Chandra Samanta. 2024. "Synthesis of an Aryl-Semicarbazone-Based Cu(II) Complex for DNA and BSA Interaction and Anti-Cancer Activity against Human Cervix Uteri Carcinoma" Inorganics 12, no. 1: 19. https://doi.org/10.3390/inorganics12010019
APA StyleMaity, R., Manna, B., Maity, S., Jana, K., Maity, T., Afzal, M., Sepay, N., & Samanta, B. C. (2024). Synthesis of an Aryl-Semicarbazone-Based Cu(II) Complex for DNA and BSA Interaction and Anti-Cancer Activity against Human Cervix Uteri Carcinoma. Inorganics, 12(1), 19. https://doi.org/10.3390/inorganics12010019