First Attempt to Couple Proteomics with the AhR Reporter Gene Bioassay in Soil Pollution Monitoring and Assessment
Abstract
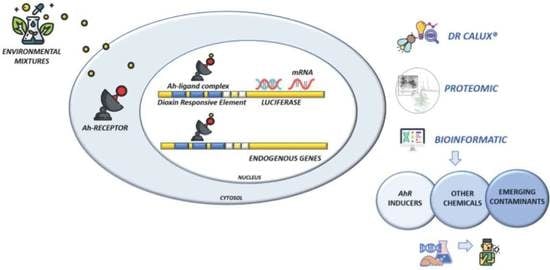
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Experimental Design
2.3. Rat Hepatoma Cell (H4IIE) Preparation for Proteomic Analysis
2.4. High-Resolution 2D Electrophoresis
2.5. Mass Spectrometry by MALDI ToF-ToF
2.6. PCA and Heatmap Analysis
2.7. Enrichment Analyses
2.7.1. Gene Ontology Terms by DAVID
2.7.2. Enrichr
2.7.3. UniProt BLAST for Human Proteins Similarity
2.7.4. Disease (by Biomarkers) Analysis by MetaCore
2.8. Chemical Analysis of Topsoil Samples
3. Results
3.1. DR-CALUX® Bioassay and GC-MS/MS Analysis of Topsoil Extracts
3.2. Proteomics Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Behnisch, P.A.; Hosoe, K.; Sakai, S. Bioanalytical screening methods for dioxins and dioxin-like compounds a review of bioassay/biomarker technology. Environ. Int. 2001, 27, 413–439. [Google Scholar] [CrossRef]
- Behnisch, P.A.; Hosoe, K.; Sakai, S. Combinatorial bio/chemical analysis of dioxin and dioxin-like compounds in waste recycling, feed/food, humans/wildlife and the environment. Environ. Int. 2001, 27, 495–519. [Google Scholar] [CrossRef]
- van Vugt-Lussenburg, B.M.A.; Pieterse, B.; Middelhof, I.; Behnisch, P.A.; van der Burg, B.; Brouwer, B. The “dirty dozend” pops & other pollutants: Toxicological profiling by calux panel. Organohalogen. Compd. 2014, 76, 3. [Google Scholar]
- Behnisch, P.A.; Hosoe, K.; Sakai, S. Brominated dioxin-like compounds: In vitro assessment in comparison to classical dioxin-like compounds and other polyaromatic compounds. Environ. Int. 2003, 29, 861–877. [Google Scholar] [CrossRef]
- Suzuki, G.; Someya, M.; Matsukami, H.; Tue, N.M.; Uchida, N.; Tuyen, L.H.; Viet, P.H.; Takahashi, S.; Tanabe, S.; Brouwer, A.; et al. Comprehensive evaluation of dioxins and dioxin-like compounds in surface soils and river sediments from e-waste-processing sites in a village in Northern Vietnam: Heading towards the environmentally sound management of e-waste. Emerg. Contam. 2016, 2, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Behnisch, P.A.; Umlauf, G.; Stachel, B.; Felzel, E.; Brouwer, B. Bio/chemical analyis of sediments from the Elbe river, the North Sea and from several tributaries. Organohalogen. Compd. 2010, 72, 5. [Google Scholar]
- Besselink, H.T.; Schipper, C.; Klamer, H.; Leonards, P.; Verhaar, H.; Felzel, E.; Murk, A.J.; Thain, J.; Hosoe, K.; Schoeters, G.; et al. Intra- and interlaboratory calibration of the DR CALUX bioassay for the analysis of dioxins and dioxin-like chemicals in sediments. Environ. Toxicol. Chem. 2004, 23, 2781–2789. [Google Scholar] [CrossRef] [Green Version]
- Budin, C.; Petrlik, J.; Strakova, J.; Hamm, S.; Beeler, B.; Behnisch, P.; Besselink, H.; van der Burg, B.; Brouwer, A. Detection of High PBDD/Fs levels and dioxin-like activity in toys using a combination of GC-HRMS, rat-based and human-based DR CALUX® reporter gene assays. Chemosphere 2020, 251, 126579. [Google Scholar] [CrossRef]
- Escher, B.I.; Allinson, M.; Altenburger, R.; Bain, P.A.; Balaguer, P.; Busch, W.; Crago, J.; Denslow, N.D.; Dopp, E.; Hilscherova, K.; et al. Benchmarking organic micropollutants in wastewater, recycled water and drinking water with in vitro bioassays. Environ. Sci. Technol. 2014, 48, 1940–1956. [Google Scholar] [CrossRef]
- Wernersson, A.-S.; Carere, M.; Maggi, C.; Tusil, P.; Soldan, P.; James, A.; Sanchez, W.; Dulio, V.; Broeg, K.; Reifferscheid, G.; et al. The European technical report on aquatic effect-based monitoring tools under the water framework directive. Environ. Sci. Eur. 2015, 27, 7. [Google Scholar] [CrossRef] [Green Version]
- Saracini, C. ECHA-Relazione 2017 Regolamento REACH. Available online: https://www.certifico.com/chemicals/documenti-chemicals/94-documenti-echa/5704-echa-relazione-2017-regolamento-reach (accessed on 9 September 2020).
- Connon, R.E.; Geist, J.; Werner, I. Effect-based tools for monitoring and predicting the ecotoxicological effects of chemicals in the aquatic environment. Sensors 2012, 12, 12741–12771. [Google Scholar] [CrossRef] [Green Version]
- Carleo, A.; Landi, C.; Prasse, A.; Bergantini, L.; D’Alessandro, M.; Cameli, P.; Janciauskiene, S.; Rottoli, P.; Bini, L.; Bargagli, E. Proteomic characterization of idiopathic pulmonary fibrosis patients: Stable versus acute exacerbation. Monaldi Arch. Chest. Dis. 2020, 90, 180–191. [Google Scholar] [CrossRef]
- Della Torre, C.; Maggioni, D.; Ghilardi, A.; Parolini, M.; Santo, N.; Landi, C.; Madaschi, L.; Magni, S.; Tasselli, S.; Ascagni, M.; et al. The interactions of fullerene C60 and benzo(α)pyrene influence their bioavailability and toxicity to zebrafish embryos. Environ. Pollut. 2018, 241, 999–1008. [Google Scholar] [CrossRef]
- Grassi, G.; Landi, C.; Torre, C.D.; Bergami, E.; Bini, L.; Corsi, I. Proteomic profile of the hard corona of charged polystyrene nanoparticles exposed to sea urchin Paracentrotus Lividus coelomic fluid highlights potential drivers of toxicity. Environ. Sci. Nano 2019, 6, 2937–2947. [Google Scholar] [CrossRef]
- Landi, C.; Carleo, A.; Vantaggiato, L.; Bergantini, L.; d’Alessandro, M.; Cameli, P.; Sebastiani, G.; Dotta, F.; Bargagli, E. Common molecular pathways targeted by nintedanib in cancer and IPF: A bioinformatic study. Pulm. Pharmacol. Ther. 2020, 64, 101941. [Google Scholar] [CrossRef]
- Ontañon, O.M.; Landi, C.; Carleo, A.; Gagliardi, A.; Bianchi, L.; González, P.S.; Agostini, E.; Bini, L. What makes A. Guillouiae SFC 500-1A able to co-metabolize phenol and Cr(VI)? A proteomic approach. J. Hazard. Mater. 2018, 354, 215–224. [Google Scholar] [CrossRef]
- Kumari, M.; Kumar, A. Identification of component-based approach for prediction of joint chemical mixture toxicity risk assessment with respect to human health: A critical review. Food Chem. Toxicol. 2020, 143, 111458. [Google Scholar] [CrossRef]
- Heys, K.A.; Shore, R.F.; Pereira, M.G.; Jones, K.C.; Martin, F.L. Risk assessment of environmental mixture effects. RSC Adv. 2016, 6, 47844–47857. [Google Scholar] [CrossRef] [Green Version]
- König, M.; Escher, B.I.; Neale, P.A.; Krauss, M.; Hilscherová, K.; Novák, J.; Teodorović, I.; Schulze, T.; Seidensticker, S.; Kamal Hashmi, M.A.; et al. Impact of untreated wastewater on a major European river evaluated with a combination of in vitro bioassays and chemical analysis. Environ. Pollut. 2017, 220, 1220–1230. [Google Scholar] [CrossRef]
- Kortenkamp, A.; Faust, M. Regulate to reduce chemical mixture risk. Science 2018, 361, 224–226. [Google Scholar] [CrossRef] [Green Version]
- Escher, B.I.; Stapleton, H.M.; Schymanski, E.L. Tracking complex mixtures of chemicals in our changing environment. Science 2020, 367, 388–392. [Google Scholar] [CrossRef]
- Drakvik, E.; Altenburger, R.; Aoki, Y.; Backhaus, T.; Bahadori, T.; Barouki, R.; Brack, W.; Cronin, M.T.D.; Demeneix, B.; Hougaard Bennekou, S.; et al. Statement on advancing the assessment of chemical mixtures and their risks for human health and the environment. Environ. Int. 2020, 134, 105267. [Google Scholar] [CrossRef]
- Altenburger, R.; Scholze, M.; Busch, W.; Escher, B.I.; Jakobs, G.; Krauss, M.; Krüger, J.; Neale, P.A.; Ait-Aissa, S.; Almeida, A.C.; et al. Mixture effects in samples of multiple contaminants—An inter-laboratory study with manifold bioassays. Environ. Int. 2018, 114, 95–106. [Google Scholar] [CrossRef]
- Escher, B.I.; Aït-Aïssa, S.; Behnisch, P.A.; Brack, W.; Brion, F.; Brouwer, A.; Buchinger, S.; Crawford, S.E.; Du Pasquier, D.; Hamers, T.; et al. Effect-based trigger values for in vitro and in vivo bioassays performed on surface water extracts supporting the environmental quality standards (EQS) of the European water framework directive. Sci. Total Environ. 2018, 628–629, 748–765. [Google Scholar] [CrossRef]
- Neale, P.A.; Altenburger, R.; Aït-Aïssa, S.; Brion, F.; Busch, W.; de Aragão Umbuzeiro, G.; Denison, M.S.; Du Pasquier, D.; Hilscherová, K.; Hollert, H.; et al. Development of a bioanalytical test battery for water quality monitoring: Fingerprinting identified micropollutants and their contribution to effects in surface water. Water Res. 2017, 123, 734–750. [Google Scholar] [CrossRef] [Green Version]
- Van der Oost, R.; Sileno, G.; Suárez-Muñoz, M.; Nguyen, M.T.; Besselink, H.; Brouwer, A. SIMONI (Smart Integrated Monitoring) as a novel bioanalytical strategy for water quality assessment: Part i-model design and effect-based trigger values. Environ. Toxicol. Chem. 2017, 36, 2385–2399. [Google Scholar] [CrossRef]
- Neale, P.A.; Ait-Aissa, S.; Brack, W.; Creusot, N.; Denison, M.S.; Deutschmann, B.; Hilscherová, K.; Hollert, H.; Krauss, M.; Novák, J.; et al. Linking in vitro effects and detected organic micropollutants in surface water using mixture-toxicity modeling. Environ. Sci. Technol. 2015, 49, 14614–14624. [Google Scholar] [CrossRef] [Green Version]
- Schiwy, A.; Brinkmann, M.; Thiem, I.; Guder, G.; Winkens, K.; Eichbaum, K.; Nüßer, L.; Thalmann, B.; Buchinger, S.; Reifferscheid, G.; et al. Determination of the CYP1A-inducing potential of single substances, mixtures and extracts of samples in the micro-EROD assay with H4IIE cells. Nat. Protoc. 2015, 10, 1728–1741. [Google Scholar] [CrossRef]
- Heinrich, P.; Petschick, L.L.; Northcott, G.L.; Tremblay, L.A.; Ataria, J.M.; Braunbeck, T. Assessment of cytotoxicity, genotoxicity and 7-Ethoxyresorufin-O-Deethylase (EROD) Induction in sediment extracts from New Zealand urban estuaries. Ecotoxicology 2017, 26, 211–226. [Google Scholar] [CrossRef]
- Liberatori, G.; Cotugno, P.; Sturba, L.; Vannuccini, M.L.; Capasso, G.; Velardo, R.; Besselink, H.; Massari, F.; Tursi, A.; Corbelli, V.; et al. Occurrence and spatial distribution of dioxin and dioxin-like compounds in topsoil of Taranto (Apulia, Italy) by GC-MS analysis and DR-CALUX® bioassay. Chemosphere 2021, 279, 130576. [Google Scholar] [CrossRef]
- Harlow, E.; Lane, D. Bradford Assay. CSH Protoc. 2006, 1121–1132. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Berg, M.; Birnbaum, L.S.; Denison, M.; De Vito, M.; Farland, W.; Feeley, M.; Fiedler, H.; Hakansson, H.; Hanberg, A.; Haws, L.; et al. The 2005 World Health Organization reevaluation of human and mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol. Sci. 2006, 93, 223–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, B.; Felzel, E.; Winter, R.; van der Burg, B.; Brouwer, A. PAH-CALUX, an optimized bioassay for AhR-mediated hazard identification of polycyclic aromatic hydrocarbons (PAHs) as individual compounds and in complex mixtures. Environ. Sci. Technol. 2013, 47, 11651–11659. [Google Scholar] [CrossRef] [PubMed]
- Notarnicola, B.; Tassielli, G.; Renzulli, P.A. Industrial symbiosis in the Taranto industrial district: Current level, constraints and potential new synergies. J. Clean. Prod. 2016, 122, 133–143. [Google Scholar] [CrossRef]
- Comba, P.; Pirastu, R.; Conti, S.; De Santis, M.; Iavarone, I.; Marsili, G.; Mincuzzi, A.; Minelli, G.; Manno, V.; Minerba, S.; et al. Environment and health in Taranto, southern Italy: Epidemiological studies and public health recommendations. Epidemiol. Prev. 2012, 36, 305–320. [Google Scholar]
- Mazza, A.; Piscitelli, P.; Neglia, C.; Della Rosa, G.; Iannuzzi, L. Illegal dumping of toxic waste and its effect on human health in Campania, Italy. Int. J. Environ. Res. Public Health 2015, 12, 6818–6831. [Google Scholar] [CrossRef] [Green Version]
- Zona, A.; Iavarone, I.; Buzzoni, C.; Conti, S.; Santoro, M.; Fazzo, L.; Pasetto, R.; Pirastu, R.; Bruno, C.; Ancona, C.; et al. SENTIERI: Epidemiological study of residents in national priority contaminated sites. Fifth report. Epidemiol. Prev. 2019, 43, 1–208. [Google Scholar] [CrossRef]
- Pascuzzi, S.; Russo, G.; Mugnozza, G.S.; Verdiani, G.; Lagattolla, G. Contamination of the environmental matrices in agricultural areas produced by industrial discharges: The case study of the Land of the City of Statte (Taranto, Southern Italy). Procedia Environ. Sci. 2013, 19, 671–680. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.-Y.; Lee, Y.-P.; Li, C.-P.; Chi, K.-H.; Liang, B.-W.P.; Liu, W.-Y.; Wang, C.-C.; Lin, S.; Chen, T.-C.; Yeh, K.-J.C.; et al. Combination of a fast cleanup procedure and a DR-CALUX® bioassay for dioxin surveillance in Taiwanese Soils. Int. J. Environ. Res. Public Health 2014, 11, 4886–4904. [Google Scholar] [CrossRef] [Green Version]
- Harischandra, D.S.; Ghaisas, S.; Rokad, D.; Kanthasamy, A.G. Exosomes in toxicology: Relevance to chemical exposure and pathogenesis of environmentally linked diseases. Toxicol. Sci. 2017, 158, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Erlichman, C. Tanespimycin: The opportunities and challenges of targeting heat shock protein 90. Expert Opin. Investig. Drugs 2009, 18, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.; Easley, C.; Kirkpatrick, P. Vorinostat. Nat. Rev. Drug Discov. 2007, 6, 21–22. [Google Scholar] [CrossRef]
- Bouquié, R.; Deslandes, G.; Mazaré, H.; Cogné, M.; Mahé, J.; Grégoire, M.; Jolliet, P. Cannabis and anticancer drugs: Societal usage and expected pharmacological interactions—A review. Fundam. Clin. Pharmacol. 2018, 32, 462–484. [Google Scholar] [CrossRef]
- Cecinato, A.; Balducci, C. Detection of cocaine in the airborne particles of the Italian cities Rome and Taranto. J. Sep. Sci. 2007, 30, 1930–1935. [Google Scholar] [CrossRef] [PubMed]
- Parlamento sul Fenomeno delle Tossicodipendenze in Italia. Relazione Annuale al Parlamento sul Fenomeno delle Tossicodipendenze in Italia Anno 2019 (Dati 2018); ISS Osservatorio Fumo, Alcol e Droga: Rome, Italy, 2020. [Google Scholar]
- Szewczyk, R.; Różalska, S.; Mironenka, J.; Bernat, P. atrazine biodegradation by mycoinsecticide metarhizium Robertsii: Insights into its amino acids and lipids profile. J. Environ. Manag. 2020, 262, 110304. [Google Scholar] [CrossRef]
- Ballar Kirmizibayrak, P.; Erbaykent-Tepedelen, B.; Gozen, O.; Erzurumlu, Y. Divergent modulation of proteostasis in prostate cancer. Adv. Exp. Med. Biol. 2020, 1233, 117–151. [Google Scholar] [CrossRef]
- Ramkumar, B.; Dharaskar, S.P.; Mounika, G.; Paithankar, K.; Sreedhar, A.S. Mitochondrial chaperone, TRAP1 as a potential pharmacological target to combat cancer metabolism. Mitochondrion 2020, 50, 42–50. [Google Scholar] [CrossRef]
- Ferreira, L.M.R.; Cunha-Oliveira, T.; Sobral, M.C.; Abreu, P.L.; Alpoim, M.C.; Urbano, A.M. Impact of carcinogenic chromium on the cellular response to proteotoxic stress. Int. J. Mol. Sci. 2019, 20, 4901. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.-Q.; Miao, X.-J.; Gao, J.; Zhou, Y.-H.; Ji, F.-Y.; Cheng, X.-B. The 150-KDa oxygen-regulated protein (ORP150) regulates proteinuria in diabetic nephropathy via mediating VEGF. Exp. Mol. Pathol. 2019, 110, 104255. [Google Scholar] [CrossRef]
- Li, X.; Zhang, N.-X.; Ye, H.-Y.; Song, P.-P.; Chang, W.; Chen, L.; Wang, Z.; Zhang, L.; Wang, N.-N. HYOU1 promotes cell growth and metastasis via activating PI3K/AKT signaling in epithelial ovarian cancer and predicts poor prognosis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4126–4135. [Google Scholar] [CrossRef]
- de Oliveira, K.M.H.; Garlet, G.P.; De Rossi, A.; Barreiros, D.; Queiroz, A.M.; da Silva, L.A.B.; Nelson-Filho, P.; da Silva, R.A.B. Effects of rosiglitazone on the outcome of experimental periapical lesions in mice. J. Endod. 2017, 43, 2061–2069. [Google Scholar] [CrossRef]
- Li, Y.; Yan, M.; Yang, J.; Raman, I.; Du, Y.; Min, S.; Fang, X.; Mohan, C.; Li, Q.-Z. Glutathione S-transferase Mu 2-transduced mesenchymal stem cells ameliorated anti-glomerular basement membrane antibody-induced glomerulonephritis by inhibiting oxidation and inflammation. Stem. Cell Res. Ther. 2014, 5, 19. [Google Scholar] [CrossRef] [Green Version]
- Kruithof, P.D.; Lunev, S.; Aguilar Lozano, S.P.; de Assis Batista, F.; Al-Dahmani, Z.M.; Joles, J.A.; Dolga, A.M.; Groves, M.R.; van Goor, H. Unraveling the role of thiosulfate sulfurtransferase in metabolic diseases. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165716. [Google Scholar] [CrossRef]
- Li, Z.; Nesbitt, N.M.; Malone, L.E.; Gnatenko, D.V.; Wu, S.; Wang, D.; Zhu, W.; Girnun, G.D.; Bahou, W.F. Heme degradation enzyme biliverdin IXβ reductase is required for stem cell glutamine metabolism. Biochem. J. 2018, 475, 1211–1223. [Google Scholar] [CrossRef]
- Manoharan, R.; Seong, H.-A.; Ha, H. Dual roles of serine-threonine kinase receptor-associated protein (STRAP) in redox-sensitive signaling pathways related to cancer development. Oxid. Med. Cell Longev. 2018, 2018, 5241524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seong, H.-A.; Jung, H.; Ha, H. NM23-H1 tumor suppressor physically interacts with serine-threonine kinase receptor-associated protein, a transforming growth factor-beta (TGF-Beta) receptor-interacting protein, and negatively regulates TGF-Beta signaling. J. Biol. Chem. 2007, 282, 12075–12096. [Google Scholar] [CrossRef] [Green Version]
- Vander Ark, A.; Cao, J.; Li, X. TGF-β receptors: In and beyond TGF-β signaling. Cell. Signal. 2018, 52, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Giua, R.; Spartera, M.; Viviano, G.; Ziemacki, G.; Carbotti, G. Cancer risk for coke-oven workers in the Taranto steel plant. Epidemiol. Prev. 2005, 29, 42–44. [Google Scholar] [PubMed]
- Pirastu, R.; Comba, P.; Iavarone, I.; Zona, A.; Conti, S.; Minelli, G.; Manno, V.; Mincuzzi, A.; Minerba, S.; Forastiere, F.; et al. Environment and health in contaminated sites: The case of Taranto, Italy. J. Environ. Public Health 2013, 2013, 753719. [Google Scholar] [CrossRef] [Green Version]
- Pirastu, R.; Ricci, P.; Comba, P.; Bianchi, F.; Biggeri, A.; Conti, S.; Fazzo, L.; Forastiere, F.; Iavarone, I.; Martuzzi, M.; et al. SENTIERI project: Discussion and conclusions. Epidemiol. Prev. 2014, 38, 125–133. [Google Scholar]
- White, S.S.; Birnbaum, L.S. An overview of the effects of dioxins and dioxin-like compounds on vertebrates, as documented in human and ecological epidemiology. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2009, 27, 197–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ARPA Puglia. Relazione sui Dati Ambientali Dell’area di Taranto. 2008. Available online: http://www.arpa.puglia.it/c/document_library/get_file?uuid¼96dc386e-2a6d-4758-8c47-e4d15d367c70&groupId¼10125 (accessed on 1 October 2021).
- Bertollini, R.; Faberi, M.; Di Tanno, N.; Division WHOEC for E and HR. Ambiente e Salute in Italia. Roma: Centro Europeo Ambiente e Salute. 1997. Available online: https://apps.who.int/iris/handle/10665/42009 (accessed on 9 September 2020).
- Marinaccio, A.; Belli, S.; Binazzi, A.; Scarselli, A.; Massari, S.; Bruni, A.; Conversano, M.; Crosignani, P.; Minerba, A.; Zona, A.; et al. Residential proximity to industrial sites in the area of Taranto (Southern Italy). A case-control cancer incidence study. Ann. Ist. Super. Sanita 2011, 47, 192–199. [Google Scholar] [PubMed]
- Martinelli, D.; Mincuzzi, A.; Minerba, S.; Tafuri, S.; Conversano, M.; Caputi, G.; Lopalco, P.L.; Quarto, M.; Germinario, C.; Prato, R. Malignant cancer mortality in Province of Taranto (Italy). Geographic analysis in an area of high environmental risk. J. Prev. Med. Hyg. 2009, 50, 181–190. [Google Scholar] [PubMed]
- Martuzzi, M.; Mitis, F.; Biggeri, A.; Terracini, B.; Bertollini, R. Environment and health status of the population in areas with high risk of environmental crisis in Italy. Epidemiol. Prev. 2002, 26, 1–53. [Google Scholar]
- Vigotti, M.A.; Mataloni, F.; Bruni, A.; Minniti, C.; Gianicolo, E.A.L. Mortality analysis by neighbourhood in a city with high levels of industrial air pollution. Int. J. Public Health 2014, 59, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Brattoli, M.; de Gennaro, G.; Carella, G.; De Gennaro, L.; Assennato, G.; Giua, R.; Angiuli, L.; Trizio, L. Integration of automatic remote systems for olfactory annoyance detection and evaluation in the City of Taranto. Chem. Eng. Trans. 2014, 40, 19–24. [Google Scholar]



| Compound | Concentrations (ng/kg d.w.) | TEQWHO | Percentage (%) |
|---|---|---|---|
| Chlorinated dibenzo-p-dioxins | |||
| 2,3,7,8-TCDD | 3.5 | 3.5 | 5.9% |
| 1,2,3,7,8-PeCDD | 9.2 | 9.2 | 15.5% |
| 1,2,3,4,7,8-HxCDD | 4.4 | 0.44 | 0.7% |
| 1,2,3,7,8,9-HxCDD | 4.8 | 0.48 | 0.8% |
| 1,2,3,4,6,7,8-HpCDD | 25.7 | 0.257 | 0.4% |
| 1,2,3,4,6,7,8,9-OCDD | 40.2 | 0.01206 | 0% |
| Chlorinated dibenzofurans | |||
| 2,3,7,8-TCDF | 13.9 | 1.39 | 2.3% |
| 1,2,3,7,8-PeCDF | 18.9 | 0.567 | 1% |
| 2,3,4,7,8-PeCDF | 83.4 | 25.02 | 42.2% |
| 1,2,3,4,7,8-HxCDF | 50.4 | 5.04 | 8.5% |
| 1,2,3,6,7,8-HxCDF | 31.6 | 3.16 | 5.3% |
| 1,2,3,7,8,9-HxCDF | 44 | 4.4 | 7.4% |
| 2,3,4,6,7,8-HxDF | 4.6 | 0.46 | 0.8% |
| 1,2,3,4,6,7,8-HpCDF | 138.1 | 1.381 | 2.3% |
| 1,2,3,4,7,8,9-HpCDF | 5.5 | 0.055 | 0.1% |
| 1,2,3,4,6,7,8,9-OCDF | 219 | 0.0657 | 0.1% |
| Non-ortho–substituted PCBs | |||
| 3,3’,4,4’-tetraCB (PCB-77) | 16 | 0 | 0% |
| 3,4,4’,5-tetraCB (PCB-81) | 7 | 0 | 0% |
| 3,3’,4,4’,5-pentaCB (PCB-126) | 37 | 3.7 | 6.2% |
| 3,3’,4,4’,5,5’-hexaCB (PCB-169) | 5 | 0.15 | 0.3% |
| Mono-ortho–substituted PCBs | |||
| 2,3,3’,4,4’-pentaCB (PCB 105) | 47 | 0 | 0% |
| 2,3,4,4’,5-pentaCB (PCB 114) | 7 | 0 | 0% |
| 2,3’,4,4’,5-pentaCB (PCB 118) | 67 | 0 | 0% |
| 2’,3,4,4’,5-pentaCB (PCB 123) | 18 | 0 | 0% |
| 2,3,3’,4,4’,5-hexaCB (PCB 156) | 45 | 0 | 0% |
| 2,3,3’,4,4’,5’-hexaCB (PCB 157) | 10 | 0 | 0% |
| 2,3’,4,4’,5,5’-hexaCB (PCB 167) | 44 | 0 | 0% |
| 2,3,3’,4,4’,5,5’-heptaCB (PCB 189) | 11 | 0 | 0% |
| average 59 | |||
| std. dev. 17 | |||
| Compound | Concentrations (mg/kg d.w.) | EC50 DR CALUX® | TEQ | Percentage (%) |
|---|---|---|---|---|
| Polycyclic Aromatic Hydrocarbons | ||||
| Pirene | 0.003 | 0% | ||
| Benzo[a]antracene | 0.00011 | 0% | ||
| Crisene | 0.004 | 0% | ||
| Benzo[b]fluorantene | 0.004 | 0.00092 | 0.00000368 | 69.4% |
| Benzo[k]fluorantene | 0.003 | 0.00054 | 0.00000162 | 30.6% |
| Benzo[a]pirene | 0.00025 | - | 0% | |
| Benzo[g,h,i]perilene | 0.003 | 0% | ||
| Dibenzo[a,h]antracene | 0.0011 | 0% | ||
| Indeno[123-c,d]antracene | - | 0.00076 | 0% | |
| Dibenzo[a,e]perilene | - | 0% | ||
| Dibenzo[a,i]perilene | - | 0% | ||
| Dibenzo[a,l]perilene | - | 0% | ||
| Dibenzo[a,h]perilene | - | 0% | ||
| ∑PAHs | 0.017 | 0.0000053 | ||
| Biological Processes Terms | % | p-Value | Benjamini |
|---|---|---|---|
| cellular response to chemical stimulus | 46.2 | 1.8 × 10−5 | 1.8 × 10−2 |
| cellular response to stress | 38.5 | 3.1 × 10−5 | 1.5 × 10−2 |
| response to inorganic substance | 26.9 | 5.7 × 10−5 | 1.9 × 10−2 |
| response to oxygen-containing compound | 38.5 | 6 × 10−5 | 1.5 × 10−2 |
| response to chemical | 57.7 | 1.2 × 10−4 | 2.4 × 10−2 |
| response to stress | 46.2 | 2.8 × 10−4 | 4.5 × 10−2 |
| response to endogenous stimulus | 34.6 | 3.9 × 10−4 | 5.3 × 10−2 |
| regulation of translation | 19.2 | 4 × 10−4 | 4.7 × 10−2 |
| regulation of apoptotic process | 30.8 | 5.2 × 10−4 | 5.6 × 10−2 |
| regulation of cellular amide metabolic process | 19.2 | 5.2 × 10−4 | 5 × 10−2 |
| regulation of programmed cell death | 30.8 | 5.6 × 10−4 | 4.9 × 10−2 |
| posttranscriptional regulation of gene expression | 19.2 | 7.5 × 10−4 | 6 × 10−2 |
| regulation of cell death | 30.8 | 9.4 × 10−4 | 6.9 × 10−2 |
| organonitrogen compound metabolic process | 34.6 | 9.8 × 10−4 | 6.7 × 10−2 |
| Cellular Component Terms | |||
| extracellular exosome | 60 | 8.2 × 10−9 | 1.4 × 10−7 |
| extracellular vesicle | 60 | 8.8 × 10−9 | 7.6 × 10−7 |
| extracellular organelle | 60 | 9 × 10−9 | 5.2 × 10−7 |
| membrane-bounded vesicle | 64 | 1.2 × 10−8 | 5 × 10−7 |
| vesicle | 64 | 2.3 × 10−8 | 8 × 10−7 |
| cytoplasmic part | 76 | 2.4 × 10−7 | 6.9 × 10−6 |
| cytosol | 48 | 5.7 × 10−7 | 1.4 × 10−5 |
| extracellular region part | 60 | 6.8 × 10−7 | 1.5 × 10−5 |
| extracellular region | 60 | 2.6 × 10−6 | 4.9 × 10−5 |
| cytoplasm | 80 | 5.3 × 10−6 | 9.1 × 10−5 |
| intracellular part | 80 | 7 × 10−4 | 1.1 × 10−2 |
| intracellular organelle | 76 | 7.4 × 10−4 | 1.1 × 10−2 |
| Molecular Function Terms | |||
| RNA binding | 36 | 2.8 × 10−4 | 5 × 10−2 |
| purine ribonucleoside triphosphate binding | 36 | 3.9 × 10−4 | 3.5 × 10−2 |
| purine ribonucleoside binding | 36 | 4 × 10−4 | 2.4 × 10−2 |
| ribonucleoside binding | 36 | 4.1 × 10−4 | 1.9 × 10−2 |
| purine nucleoside binding | 36 | 4.1 × 10−4 | 1.9 × 10−2 |
| nucleoside binding | 36 | 4.2 × 10−4 | 1.5 × 10−2 |
| purine ribonucleotide binding | 36 | 4.6 × 10−4 | 1.4 × 10−2 |
| purine nucleotide binding | 36 | 4.8 × 10−4 | 1.3 × 10−2 |
| ribonucleotide binding | 36 | 4.9 × 10−4 | 1.1 × 10−2 |
| ATP binding | 32 | 6 × 10−4 | 1.2 × 10−2 |
| adenyl ribonucleotide binding | 32 | 7.1 × 10−4 | 1.3 × 10−2 |
| adenyl nucleotide binding | 32 | 7.4 × 10−4 | 1.2 × 10−2 |
| small molecule binding | 40 | 8.6 × 10−4 | 1.3 × 10−2 |
| Term | p-Value | Genes |
|---|---|---|
| (17S)-17-hydroxy-13,17-dimethyl-1,2,6,7,8,14,15,16-octahydrocyclopenta[a]phenanthren-3-one CTD 00007088 | 1.39 × 10−7 | PDXK; PRDX1; ASNS; TALDO1; HYOU1; EEF2; PAK2; ACTG1 |
| 67526-95-8 CTD 00007263 | 1.83 × 10−10 | TRAP1; TST; MVP; G3BP1; ANXA5; ASNS; HYOU1; PFAS; ACTG1 |
| Vorinostat CTD 00003560 | 1.43 × 10−11 | TRAP1; PRDX1; ANXA5; TALDO1; STRAP; BLVRB |
| troglitazone CTD 00002415 | 1.53 × 10−12 | HSPH1; MVP; PRDX1; ANXA5; ASNS; ACTG1 |
| chlortetracycline HL60 DOWN | 2.94 × 10−12 | PDXK; HSPH1; TST; PRDX1; CRYZL1; UBA1; PAK2 |
| clonidine HL60 DOWN | 3.74 × 10−11 | HSPH1; PFAS; EIF4E |
| lobeline HL60 DOWN | 4.72 × 10−12 | PDXK; HSPH1; G3BP1; CRYZL1; TALDO1; PFAS; PAK2; EIF4E |
| PERHEXILINE CTD 00006493 | 4.79 × 10−11 | ASNS; EIF4E |
| POTASSIUM DICHROMATE CTD 00006598 | 6.61 × 10−11 | TRAP1; MVP; PRDX1; ASNS; UBA1 |
| atrazine CTD 00005450 | 6.7 × 10−11 | TRAP1; GSTM2; HSPH1; TST; MVP; PRDX1; G3BP1; ANXA5; CRYZL1; ASNS; PFAS |
| cyproheptadine PC3 UP | 7.38 × 10−11 | ASNS; HYOU1 |
| clindamycin HL60 DOWN | 7.75 × 10−11 | TRAP1; PDXK; PITRM1; PRDX1; CRYZL1; ASNS; TALDO1; PAK2 |
| Copper sulfate CTD 00007279 | 8.72 × 10−10 | TRAP1; PDXK; MVP; ANXA5; ASNS; TALDO1; ACTG1; KLC4; HSPH1; PITRM1; TST; G3BP1; CRYZL1; BLVRB; PAK2; EIF4E |
| tanespimycin SKMEL5 UP | 9.42 × 10−11 | HSPH1; ASNS |
| Term | p-Value | Genes |
|---|---|---|
| Eukaryotic protein translation | 0.002 | EEF2; EIF4E |
| TGF-beta signaling pathway | 0.002 | TRAP1; STRAP; PAK2 |
| Translation factors | 0.002 | EEF2; EIF4E |
| HIV-1 Nef as negative effector of Fas and TNF | 0.003 | PAK2; ACTG1 |
| Signaling events mediated by hepatocyte growth factor receptor (c-Met) | 0.005 | PAK2; EIF4E |
| Unfolded protein response | 0.005 | ASNS; HYOU1 |
| Sulfide oxidation to sulfate | 0.006 | TST |
| Heme degradation | 0.006 | BLVRB |
| Vitamin B6 metabolism | 0.008 | PDXK |
| eIF4E release | 0.008 | EIF4E |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landi, C.; Liberatori, G.; Cotugno, P.; Sturba, L.; Vannuccini, M.L.; Massari, F.; Miniero, D.V.; Tursi, A.; Shaba, E.; Behnisch, P.A.; et al. First Attempt to Couple Proteomics with the AhR Reporter Gene Bioassay in Soil Pollution Monitoring and Assessment. Toxics 2022, 10, 9. https://doi.org/10.3390/toxics10010009
Landi C, Liberatori G, Cotugno P, Sturba L, Vannuccini ML, Massari F, Miniero DV, Tursi A, Shaba E, Behnisch PA, et al. First Attempt to Couple Proteomics with the AhR Reporter Gene Bioassay in Soil Pollution Monitoring and Assessment. Toxics. 2022; 10(1):9. https://doi.org/10.3390/toxics10010009
Chicago/Turabian StyleLandi, Claudia, Giulia Liberatori, Pietro Cotugno, Lucrezia Sturba, Maria Luisa Vannuccini, Federica Massari, Daniela Valeria Miniero, Angelo Tursi, Enxhi Shaba, Peter A. Behnisch, and et al. 2022. "First Attempt to Couple Proteomics with the AhR Reporter Gene Bioassay in Soil Pollution Monitoring and Assessment" Toxics 10, no. 1: 9. https://doi.org/10.3390/toxics10010009
APA StyleLandi, C., Liberatori, G., Cotugno, P., Sturba, L., Vannuccini, M. L., Massari, F., Miniero, D. V., Tursi, A., Shaba, E., Behnisch, P. A., Carleo, A., Di Giuseppe, F., Angelucci, S., Bini, L., & Corsi, I. (2022). First Attempt to Couple Proteomics with the AhR Reporter Gene Bioassay in Soil Pollution Monitoring and Assessment. Toxics, 10(1), 9. https://doi.org/10.3390/toxics10010009