Proteomic Profiling of Fallopian Tube-Derived Extracellular Vesicles Using a Microfluidic Tissue-on-Chip System
Abstract
:1. Introduction
2. Materials and Methods
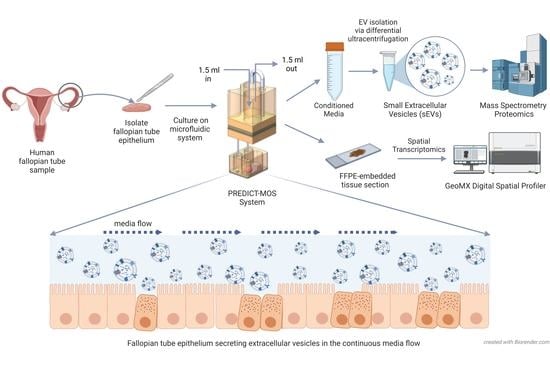
2.1. Culturing of Human Fallopian Tube (hFTE) Tissue Explants on the PREDICT-Multi-Organ-System
2.2. Live/Dead Staining
2.3. Spatial Transcriptomic Analysis to Characterize Epithelial Cell Types of Fallopian Tube Explant Cultured in the PREDICT-Multi-Organ-System
2.4. sEV Isolation and Characterization
2.5. ExoView Analysis of sEVs
2.6. Proteomics Analysis of sEV Cargos Secreted by Fallopian Tube Epithelium
2.7. Immunohistochemistry (IHC) Staining
2.8. Transmission Electron Microscopy (TEM)
2.9. Statistics
2.10. Reagents and Kits Used
3. Results
3.1. Primary Fallopian Tube Explant Maintains Epithelial Architecture and Distinct Cell Subtypes during Long-Term Culture in the Dynamic Organ-On-Chip System
3.2. Physical and Molecular Characterization of Small Extracellular Vesicles (sEVs) Derived from hFTE Tissue Explants Cultured in the PREDICT-MOS
3.3. Proteomics Profile of hFTE sEVs
3.4. The Comparison of Fallopian Tube sEV Protein Content across Different Species and with Benign Fallopian Tube and STIC Lesions in the Tissue
3.5. Correlation of hFTE sEV Proteomics Profile with Cancer-Associated hFTE Tissue Explant Transcriptome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, S.; Winuthayanon, W. Oviduct: Roles in Fertilization and Early Embryo Development. J. Endocrinol. 2017, 232, R1–R26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almiñana, C.; Heath, P.R.; Wilkinson, S.; Sanchez-Osorio, J.; Cuello, C.; Parrilla, I.; Gil, M.A.; Vazquez, J.L.; Vazquez, J.M.; Roca, J.; et al. Early Developing Pig Embryos Mediate Their Own Environment in the Maternal Tract. PLoS ONE 2012, 7, e33625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almiñana, C.; Corbin, E.; Tsikis, G.; Alcântara-Neto, A.S.; Labas, V.; Reynaud, K.; Galio, L.; Uzbekov, R.; Garanina, A.S.; Druart, X.; et al. Oviduct Extracellular Vesicles Protein Content and Their Role during Oviduct–Embryo Cross-Talk. Reproduction 2017, 154, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Winuthayanon, W.; Bernhardt, M.L.; Padilla-Banks, E.; Myers, P.H.; Edin, M.L.; Lih, F.B.; Hewitt, S.C.; Korach, K.S.; Williams, C.J. Oviductal Estrogen Receptor α Signaling Prevents Protease-Mediated Embryo Death. eLife 2015, 4, e10453. [Google Scholar] [CrossRef]
- Georgiou, A.S.; Sostaric, E.; Wong, C.H.; Snijders, A.P.L.; Wright, P.C.; Moore, H.D.; Fazeli, A. Gametes Alter the Oviductal Secretory Proteome. Mol. Cell. Proteom. 2005, 4, 1785–1796. [Google Scholar] [CrossRef] [Green Version]
- Labidi-Galy, S.I.; Papp, E.; Hallberg, D.; Niknafs, N.; Adleff, V.; Noe, M.; Bhattacharya, R.; Novak, M.; Jones, S.; Phallen, J.; et al. High Grade Serous Ovarian Carcinomas Originate in the Fallopian Tube. Nat. Commun. 2017, 8, 1093. [Google Scholar] [CrossRef] [Green Version]
- Perets, R.; Wyant, G.A.; Muto, K.W.; Bijron, J.G.; Poole, B.B.; Chin, K.T.; Chen, J.Y.H.; Ohman, A.W.; Stepule, C.D.; Kwak, S.; et al. Transformation of the Fallopian Tube Secretory Epithelium Leads to High-Grade Serous Ovarian Cancer in Brca;Tp53;Pten Models. Cancer Cell 2013, 24, 751–765. [Google Scholar] [CrossRef] [Green Version]
- Roh, M.H.; Yassin, Y.; Miron, A.; Mehra, K.K.; Mehrad, M.; Monte, N.M.; Mutter, G.L.; Nucci, M.R.; Ning, G.; Mckeon, F.D.; et al. High-Grade Fimbrial-Ovarian Carcinomas Are Unified by Altered P53, PTEN and PAX2 Expression. Mod. Pathol. 2010, 23, 1316–1324. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, L.; Wang, Y.; Tang, S.N.; Zheng, W. Fallopian Tube Secretory Cell Expansion: A Sensitive Biomarker for Ovarian Serous Carcinogenesis. Am. J. Transl. Res. 2015, 7, 2082–2090. [Google Scholar]
- Shih, I.-M.; Wang, Y.; Wang, T.-L. The Origin of Ovarian Cancer Species and Precancerous Landscape. Am. J. Pathol. 2021, 191, 26–39. [Google Scholar] [CrossRef]
- Hanley, G.E.; Pearce, C.L.; Talhouk, A.; Kwon, J.S.; Finlayson, S.J.; McAlpine, J.N.; Huntsman, D.G.; Miller, D. Outcomes from Opportunistic Salpingectomy for Ovarian Cancer Prevention. JAMA Netw. Open 2022, 5, e2147343. [Google Scholar] [CrossRef]
- Jia, Y.; Yu, L.; Ma, T.; Xu, W.; Qian, H.; Sun, Y.; Shi, H. Small Extracellular Vesicles Isolation and Separation: Current Techniques, Pending Questions and Clinical Applications. Theranostics 2022, 12, 6548–6575. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [Green Version]
- Atay, S.; Banskota, S.; Crow, J.; Sethi, G.; Rink, L.; Godwin, A.K. Oncogenic KIT-Containing Exosomes Increase Gastrointestinal Stromal Tumor Cell Invasion. Proc. Natl. Acad. Sci. USA 2014, 111, 711–716. [Google Scholar] [CrossRef] [Green Version]
- Atay, S.; Godwin, A.K. Tumor-Derived Exosomes: A Message Delivery System for Tumor Progression. Commun. Integr. Biol. 2014, 7, e28231. [Google Scholar] [CrossRef]
- Crow, J.; Atay, S.; Banskota, S.; Artale, B.; Schmitt, S.; Godwin, A.K. Exosomes as Mediators of Platinum Resistance in Ovarian Cancer. Oncotarget 2017, 8, 11917–11936. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Aranda, E.; Hayakawa, Y.; Bhanja, P.; Atay, S.; Brodin, N.P.; Li, J.; Asfaha, S.; Liu, L.; Tailor, Y.; et al. Macrophage-Derived Extracellular Vesicle-Packaged WNTs Rescue Intestinal Stem Cells and Enhance Survival after Radiation Injury. Nat. Commun. 2016, 7, 13096. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Soder, R.; Abhyankar, S.; Abdelhakim, H.; Braun, M.W.; Trinidad, C.V.; Pathak, H.B.; Pessetto, Z.; Deighan, C.; Ganguly, S.; et al. WJMSC-Derived Small Extracellular Vesicle Enhance T Cell Suppression through PD-L1. J. Extracell. Vesicles 2021, 10, e12067. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Sherman-Samis, M.; Onallah, H.; Holth, A.; Reich, R.; Davidson, B. SOX2 and SOX9 Are Markers of Clinically Aggressive Disease in Metastatic High-Grade Serous Carcinoma. Gynecol. Oncol. 2019, 153, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.A.; Stephens, K.K.; Winuthayanon, W. Extracellular Vesicles and the Oviduct Function. Int. J. Mol. Sci. 2020, 21, 8280. [Google Scholar] [CrossRef] [PubMed]
- Lange-Consiglio, A.; Perrini, C.; Albini, G.; Modina, S.; Lodde, V.; Orsini, E.; Esposti, P.; Cremonesi, F. Oviductal Microvesicles and Their Effect on in Vitro Maturation of Canine Oocytes. Reproduction 2017, 154, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Alcântara-Neto, A.S.; Schmaltz, L.; Caldas, E.; Blache, M.-C.; Mermillod, P.; Almiñana, C. Porcine Oviductal Extracellular Vesicles Interact with Gametes and Regulate Sperm Motility and Survival. Theriogenology 2020, 155, 240–255. [Google Scholar] [CrossRef]
- Al-Dossary, A.A.; Strehler, E.E.; Martin-DeLeon, P.A. Expression and Secretion of Plasma Membrane Ca2+-ATPase 4a (PMCA4a) during Murine Estrus: Association with Oviductal Exosomes and Uptake in Sperm. PLoS ONE 2013, 8, e80181. [Google Scholar] [CrossRef] [Green Version]
- Bathala, P.; Fereshteh, Z.; Li, K.; Al-Dossary, A.A.; Galileo, D.S.; Martin-DeLeon, P.A. Oviductal Extracellular Vesicles (Oviductosomes, OVS) Are Conserved in Humans: Murine OVS Play a Pivotal Role in Sperm Capacitation and Fertility. Mol. Hum. Reprod. 2018, 24, 143–157. [Google Scholar] [CrossRef] [Green Version]
- Russo, A.; Yang, Z.; Heyrman, G.M.; Cain, B.P.; Lopez Carrero, A.; Isenberg, B.C.; Dean, M.J.; Coppeta, J.; Burdette, J.E. Versican Secreted by the Ovary Links Ovulation and Migration in Fallopian Tube Derived Serous Cancer. Cancer Lett. 2022, 543, 215779. [Google Scholar] [CrossRef]
- Jackson-Bey, T.; Colina, J.; Isenberg, B.C.; Coppeta, J.; Urbanek, M.; Kim, J.J.; Woodruff, T.K.; Burdette, J.E.; Russo, A. Exposure of Human Fallopian Tube Epithelium to Elevated Testosterone Results in Alteration of Cilia Gene Expression and Beating. Hum. Reprod. 2020, 35, 2086–2096. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Ferraz, M.A.M.M.; Rho, H.S.; Hemerich, D.; Henning, H.H.W.; van Tol, H.T.A.; Hölker, M.; Besenfelder, U.; Mokry, M.; Vos, P.L.A.M.; Stout, T.A.E.; et al. An Oviduct-on-a-Chip Provides an Enhanced in Vitro Environment for Zygote Genome Reprogramming. Nat. Commun. 2018, 9, 4934. [Google Scholar] [CrossRef] [Green Version]
- Mah, L.-J.; El-Osta, A.; Karagiannis, T.C. ΓH2AX: A Sensitive Molecular Marker of DNA Damage and Repair. Leukemia 2010, 24, 679–686. [Google Scholar] [CrossRef] [Green Version]
- Ferraz, M.D.A.; Carothers, A.; Dahal, R.; Noonan, M.J.; Songsasen, N. Oviductal Extracellular Vesicles Interact with the Spermatozoon’s Head and Mid-Piece and Improves Its Motility and Fertilizing Ability in the Domestic Cat. Sci. Rep. 2019, 9, 9484. [Google Scholar] [CrossRef] [Green Version]
- Laezer, I.; Palma-Vera, S.E.; Liu, F.; Frank, M.; Trakooljul, N.; Vernunft, A.; Schoen, J.; Chen, S. Dynamic Profile of EVs in Porcine Oviductal Fluid during the Periovulatory Period. Reproduction 2020, 159, 371–382. [Google Scholar] [CrossRef]
- Acland, M.; Arentz, G.; Mussared, M.; Whitehead, F.; Hoffmann, P.; Klingler-Hoffmann, M.; Oehler, M.K. Proteomic Analysis of Pre-Invasive Serous Lesions of the Endometrium and Fallopian Tube Reveals Their Metastatic Potential. Front. Oncol. 2020, 10, 523989. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, L. The Role of ALDH2 in Tumorigenesis and Tumor Progression: Targeting ALDH2 as a Potential Cancer Treatment. Acta Pharm. Sin. B 2021, 11, 1400–1411. [Google Scholar] [CrossRef]
- Boudhraa, Z.; Carmona, E.; Provencher, D.; Mes-Masson, A.-M. Ran GTPase: A Key Player in Tumor Progression and Metastasis. Front. Cell Dev. Biol. 2020, 8, 345. [Google Scholar] [CrossRef]
- Wang, P.; Zeng, Y.; Liu, T.; Zhang, C.; Yu, P.-W.; Hao, Y.-X.; Luo, H.-X.; Liu, G. Chloride Intracellular Channel 1 Regulates Colon Cancer Cell Migration and Invasion through ROS/ERK Pathway. World J. Gastroenterol. 2014, 20, 2071–2078. [Google Scholar] [CrossRef]
- Moudry, P.; Lukas, C.; Macurek, L.; Hanzlikova, H.; Hodny, Z.; Lukas, J.; Bartek, J. Ubiquitin-Activating Enzyme UBA1 Is Required for Cellular Response to DNA Damage. Cell Cycle 2012, 11, 1573–1582. [Google Scholar] [CrossRef]
- Skornicka, E.L.; Kiyatkina, N.; Weber, M.C.; Tykocinski, M.L.; Koo, P.H. Pregnancy Zone Protein Is a Carrier and Modulator of Placental Protein-14 in T-Cell Growth and Cytokine Production. Cell. Immunol. 2004, 232, 144–156. [Google Scholar] [CrossRef]
- Cater, J.H.; Kumita, J.R.; Zeineddine Abdallah, R.; Zhao, G.; Bernardo-Gancedo, A.; Henry, A.; Winata, W.; Chi, M.; Grenyer, B.S.F.; Townsend, M.L.; et al. Human Pregnancy Zone Protein Stabilizes Misfolded Proteins Including Preeclampsia- and Alzheimer’s-Associated Amyloid Beta Peptide. Proc. Natl. Acad. Sci. USA 2019, 116, 6101–6110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witke, W.; Sutherland, J.D.; Sharpe, A.; Arai, M.; Kwiatkowski, D.J. Profilin I Is Essential for Cell Survival and Cell Division in Early Mouse Development. Proc. Natl. Acad. Sci. USA 2001, 98, 3832–3836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Tian, L.; Chi, C.; Wu, X.; Yang, X.; Han, M.; Xu, T.; Zhuang, Y.; Deng, K. Adam10 Is Essential for Early Embryonic Cardiovascular Development. Dev. Dyn. 2010, 239, 2594–2602. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, M.; Etkovitz, N.; Breitbart, H. Role and Regulation of Sperm Gelsolin Prior to Fertilization. J. Biol. Chem. 2010, 285, 39702–39709. [Google Scholar] [CrossRef] [Green Version]
- Miyado, K.; Yamada, G.; Yamada, S.; Hasuwa, H.; Nakamura, Y.; Ryu, F.; Suzuki, K.; Kosai, K.; Inoue, K.; Ogura, A.; et al. Requirement of CD9 on the Egg Plasma Membrane for Fertilization. Science 2000, 287, 321–324. [Google Scholar] [CrossRef]
- Feng, H.L.; Sandlow, J.I.; Sparks, A.E.T. Decreased Expression of the Heat Shock Protein Hsp70-2 Is Associated with the Pathogenesis of Male Infertility. Fertil. Steril. 2001, 76, 1136–1139. [Google Scholar] [CrossRef]
- Sun, P.; Wang, Y.; Gao, T.; Li, K.; Zheng, D.; Liu, A.; Ni, Y. Hsp90 Modulates Human Sperm Capacitation via the Erk1/2 and P38 MAPK Signaling Pathways. Reprod. Biol. Endocrinol. 2021, 19, 39. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, Z.; Peng, H.; Ward, S.M.; Hennig, G.W.; Zheng, H.; Yan, W. Oviductal Motile Cilia Are Essential for Oocyte Pickup but Dispensable for Sperm and Embryo Transport. Proc. Natl. Acad. Sci. USA 2021, 118, e2102940118. [Google Scholar] [CrossRef]
- Mirkin, S.; Arslan, M.; Churikov, D.; Corica, A.; Diaz, J.I.; Williams, S.; Bocca, S.; Oehninger, S. In Search of Candidate Genes Critically Expressed in the Human Endometrium during the Window of Implantation. Hum. Reprod. 2005, 20, 2104–2117. [Google Scholar] [CrossRef]
- Garrido-Gómez, T.; Dominguez, F.; Quiñonero, A.; Estella, C.; Vilella, F.; Pellicer, A.; Simon, C. Annexin A2 Is Critical for Embryo Adhesiveness to the Human Endometrium by RhoA Activation through F-actin Regulation. FASEB J. 2012, 26, 3715–3727. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Zaoui, K.; Boudhraa, Z.; Khalifé, P.; Carmona, E.; Provencher, D.; Mes-Masson, A.-M. Ran Promotes Membrane Targeting and Stabilization of RhoA to Orchestrate Ovarian Cancer Cell Invasion. Nat. Commun. 2019, 10, 2666. [Google Scholar] [CrossRef] [Green Version]
- Akinjiyan, F.A.; Dave, R.M.; Alpert, E.; Longmore, G.D.; Fuh, K.C. DDR2 Expression in Cancer-Associated Fibroblasts Promotes Ovarian Cancer Tumor Invasion and Metastasis through Periostin-ITGB1. Cancers 2022, 14, 3482. [Google Scholar] [CrossRef]
- Wei, B.-R.; Hoover, S.B.; Ross, M.M.; Zhou, W.; Meani, F.; Edwards, J.B.; Spehalski, E.I.; Risinger, J.I.; Alvord, W.G.; Quiñones, O.A.; et al. Serum S100A6 Concentration Predicts Peritoneal Tumor Burden in Mice with Epithelial Ovarian Cancer and Is Associated with Advanced Stage in Patients. PLoS ONE 2009, 4, e7670. [Google Scholar] [CrossRef]
- Yue, H.; Li, W.; Chen, R.; Wang, J.; Lu, X.; Li, J. Stromal POSTN Induced by TGF-Β1 Facilitates the Migration and Invasion of Ovarian Cancer. Gynecol. Oncol. 2021, 160, 530–538. [Google Scholar] [CrossRef]
- Jeong, K.J.; Park, S.Y.; Cho, K.H.; Sohn, J.S.; Lee, J.; Kim, Y.K.; Kang, J.; Park, C.G.; Han, J.W.; Lee, H.Y. The Rho/ROCK Pathway for Lysophosphatidic Acid-Induced Proteolytic Enzyme Expression and Ovarian Cancer Cell Invasion. Oncogene 2012, 31, 4279–4289. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Santiskulvong, C.; Rao, J.; Gimzewski, J.K.; Dorigo, O. The Role of Rho GTPase in Cell Stiffness and Cisplatin Resistance in Ovarian Cancer Cells. Integr. Biol. 2014, 6, 611–617. [Google Scholar] [CrossRef]
- Lecker, L.S.M.; Berlato, C.; Maniati, E.; Delaine-Smith, R.; Pearce, O.M.T.; Heath, O.; Nichols, S.J.; Trevisan, C.; Novak, M.; McDermott, J.; et al. TGFBI Production by Macrophages Contributes to an Immunosuppressive Microenvironment in Ovarian Cancer. Cancer Res. 2021, 81, 5706–5719. [Google Scholar] [CrossRef]
- Meng, E.; Mitra, A.; Tripathi, K.; Finan, M.A.; Scalici, J.; McClellan, S.; Madeira da Silva, L.; Reed, E.; Shevde, L.A.; Palle, K.; et al. ALDH1A1 Maintains Ovarian Cancer Stem Cell-like Properties by Altered Regulation of Cell Cycle Checkpoint and DNA Repair Network Signaling. PloS ONE 2014, 9, e107142. [Google Scholar] [CrossRef] [Green Version]
- Eskelinen, E.-L.; Illert, A.L.; Tanaka, Y.; Schwarzmann, G.; Blanz, J.; Von Figura, K.; Saftig, P. Role of LAMP-2 in Lysosome Biogenesis and Autophagy. Mol. Biol. Cell 2002, 13, 3355–3368. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Wang, J.; Wu, Y.; Wan, P.; Yang, G. Rap1A Promotes Ovarian Cancer Metastasis via Activation of ERK/P38 and Notch Signaling. Cancer Med. 2016, 5, 3544–3554. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, E. Ovarian Cancer Development and Metastasis. Am. J. Pathol. 2010, 177, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; Lin, W.; Wang, Y.; Zhang, J.; Chambers, S.K.; Li, B.; Lea, J.; Wang, Y.; Wang, Y.; Zheng, W. Loss of Tubal Ciliated Cells as a Risk for “Ovarian” or Pelvic Serous Carcinoma. Am. J. Cancer Res. 2020, 10, 3815–3827. [Google Scholar] [PubMed]
- Guo, Y.; Li, M.; Bai, G.; Li, X.; Sun, Z.; Yang, J.; Wang, L.; Sun, J. Filamin A Inhibits Tumor Progression through Regulating BRCA1 Expression in Human Breast Cancer. Oncol. Lett. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Oh, H.J.; Kim, M.J.; Lee, B.C. Canine Oviductal Exosomes Improve Oocyte Development via EGFR/MAPK Signaling Pathway. Reproduction 2020, 160, 613–625. [Google Scholar] [CrossRef]
- Kwon, J.; Jeong, S.; Choi, I.; Kim, N.-H. ADAM10 Is Involved in Cell Junction Assembly in Early Porcine Embryo Development. PLoS ONE 2016, 11, e0152921. [Google Scholar] [CrossRef] [Green Version]
- Navarathna, D.H.M.L.P.; Stein, E.V.; Lessey-Morillon, E.C.; Nayak, D.; Martin-Manso, G.; Roberts, D.D. CD47 Promotes Protective Innate and Adaptive Immunity in a Mouse Model of Disseminated Candidiasis. PLoS ONE 2015, 10, e0128220. [Google Scholar] [CrossRef] [Green Version]
- d’Almeida, T.C.; Sadissou, I.; Sagbohan, M.; Milet, J.; Avokpaho, E.; Gineau, L.; Sabbagh, A.; Moutairou, K.; Donadi, E.A.; Favier, B.; et al. High Level of Soluble Human Leukocyte Antigen (HLA)-G at Beginning of Pregnancy as Predictor of Risk of Malaria during Infancy. Sci. Rep. 2019, 9, 9160. [Google Scholar] [CrossRef] [Green Version]
- Gordon, L.I.; Douglas, S.D.; Kay, N.E.; Yamada, O.; Osserman, E.F.; Jacob, H.S. Modulation of Neutrophil Function by Lysozyme. Potential Negative Feedback System of Inflammation. J. Clin. Investig. 1979, 64, 226–232. [Google Scholar] [CrossRef] [Green Version]
- Metzler, K.D.; Fuchs, T.A.; Nauseef, W.M.; Reumaux, D.; Roesler, J.; Schulze, I.; Wahn, V.; Papayannopoulos, V.; Zychlinsky, A. Myeloperoxidase Is Required for Neutrophil Extracellular Trap Formation: Implications for Innate Immunity. Blood 2011, 117, 953–959. [Google Scholar] [CrossRef] [Green Version]
- Binotti, B.; Jahn, R.; Chua, J.J.E. Functions of Rab Proteins at Presynaptic Sites. Cells 2016, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Zhou, X.; Su, P.; Li, H.; Tu, Y.; Du, J.; Pan, C.; Wei, X.; Zheng, M.; Jin, K.; et al. Breast Cancer Cell-Derived Extracellular Vesicles Promote CD8+ T Cell Exhaustion via TGF-β Type II Receptor Signaling. Nat. Commun. 2022, 13, 4461. [Google Scholar] [CrossRef]
- Nigri, J.; Leca, J.; Tubiana, S.-S.; Finetti, P.; Guillaumond, F.; Martinez, S.; Lac, S.; Iovanna, J.L.; Audebert, S.; Camoin, L.; et al. CD9 Mediates the Uptake of Extracellular Vesicles from Cancer-Associated Fibroblasts That Promote Pancreatic Cancer Cell Aggressiveness. Sci. Signal. 2022, 15, eabg8191. [Google Scholar] [CrossRef]
- Tian, W.; Lei, N.; Zhou, J.; Chen, M.; Guo, R.; Qin, B.; Li, Y.; Chang, L. Extracellular Vesicles in Ovarian Cancer Chemoresistance, Metastasis, and Immune Evasion. Cell Death Dis. 2022, 13, 1–12. [Google Scholar] [CrossRef]
- Smith, J.M.; Wira, C.R.; Fanger, M.W.; Shen, L. Human Fallopian Tube Neutrophils–A Distinct Phenotype from Blood Neutrophils. Am. J. Reprod. Immunol. 2006, 56, 218–229. [Google Scholar] [CrossRef]
- Givan, A.L.; White, H.D.; Stern, J.E.; Colby, E.; Guyre, P.M.; Wira, C.R.; Gosselin, E.J. Flow Cytometric Analysis of Leukocytes in the Human Female Reproductive Tract: Comparison of Fallopian Tube, Uterus, Cervix, and Vagina. Am. J. Reprod. Immunol. 1997, 38, 350–359. [Google Scholar] [CrossRef]
- Lin, A.; Yan, W.-H. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front. Immunol. 2018, 9, 2164. [Google Scholar] [CrossRef]
- Fanger, N.A.; Liu, C.; Guyre, P.M.; Wardwell, K.; O’Neil, J.; Guo, T.L.; Christian, T.P.; Mudzinski, S.P.; Gosselin, E.J. Activation of Human T Cells by Major Histocompatability Complex Class II Expressing Neutrophils: Proliferation in the Presence of Superantigen, But Not Tetanus Toxoid. Blood 1997, 89, 4128–4135. [Google Scholar] [CrossRef]
- Reinisch, W.; Lichtenberger, C.; Steger, G.; Tillinger, W.; Scheiner, O.; Gangl, A.; Maurer, D.; Willheim, M. Donor Dependent, Interferon-Gamma Induced HLA-DR Expression on Human Neutrophils in Vivo. Clin. Exp. Immunol. 2003, 133, 476–484. [Google Scholar] [CrossRef]
- Naccasha, N.; Gervasi, M.-T.; Chaiworapongsa, T.; Berman, S.; Yoon, B.H.; Maymon, E.; Romero, R. Phenotypic and Metabolic Characteristics of Monocytes and Granulocytes in Normal Pregnancy and Maternal Infection. Am. J. Obstet. Gynecol. 2001, 185, 1118–1123. [Google Scholar] [CrossRef]
- Broxmeyer, H.E.; Smithyman, A.; Eger, R.R.; Meyers, P.A.; de Sousa, M. Identification of Lactoferrin as the Granulocyte-Derived Inhibitor of Colony-Stimulating Activity Production. J. Exp. Med. 1978, 148, 1052–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardighieri, L.; Lonardi, S.; Moratto, D.; Facchetti, F.; Shih, I.-M.; Vermi, W.; Kurman, R.J. Characterization of the Immune Cell Repertoire in the Normal Fallopian Tube. Int. J. Gynecol. Pathol. 2014, 33, 581–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavakis, T.; Bierhaus, A.; Nawroth, P. RAGE (Receptor for Advanced Glycation End Products): A Central Player in the Inflammatory Response. Microbes Infect. 2004, 6, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.M.; Yan, S.D.; Yan, S.F.; Stern, D.M. The Multiligand Receptor RAGE as a Progression Factor Amplifying Immune and Inflammatory Responses. J. Clin. Investig. 2001, 108, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Abu, N.; Rus Bakarurraini, N.A.A.; Nasir, S.N. Extracellular Vesicles and DAMPs in Cancer: A Mini-Review. Front. Immunol. 2021, 12, 740548. [Google Scholar] [CrossRef]
- Han, N.; Heublein, S.; Jeschke, U.; Kuhn, C.; Hester, A.; Czogalla, B.; Mahner, S.; Rottmann, M.; Mayr, D.; Schmoeckel, E.; et al. The G-Protein-Coupled Estrogen Receptor (GPER) Regulates Trimethylation of Histone H3 at Lysine 4 and Represses Migration and Proliferation of Ovarian Cancer Cells In Vitro. Cells 2021, 10, 619. [Google Scholar] [CrossRef]
- Ignatov, T.; Modl, S.; Thulig, M.; Weißenborn, C.; Treeck, O.; Ortmann, O.; Zenclussen, A.; Costa, S.; Kalinski, T.; Ignatov, A. GPER-1 Acts as a Tumor Suppressor in Ovarian Cancer. J. Ovarian Res. 2013, 6, 51. [Google Scholar] [CrossRef] [Green Version]
- Mitra, A.K.; Sawada, K.; Tiwari, P.; Mui, K.; Gwin, K.; Lengyel, E. Ligand-Independent Activation of c-Met by Fibronectin and A5β1-Integrin Regulates Ovarian Cancer Invasion and Metastasis. Oncogene 2011, 30, 1566–1576. [Google Scholar] [CrossRef] [Green Version]
- Doberstein, K.; Spivak, R.; Reavis, H.D.; Hooda, J.; Feng, Y.; Kroeger, P.T.; Stuckelberger, S.; Mills, G.B.; Devins, K.M.; Schwartz, L.E.; et al. L1CAM Is Required for Early Dissemination of Fallopian Tube Carcinoma Precursors to the Ovary. Commun. Biol. 2022, 5, 1362. [Google Scholar] [CrossRef]
- Almiñana, C.; Tsikis, G.; Labas, V.; Uzbekov, R.; da Silveira, J.C.; Bauersachs, S.; Mermillod, P. Deciphering the Oviductal Extracellular Vesicles Content across the Estrous Cycle: Implications for the Gametes-Oviduct Interactions and the Environment of the Potential Embryo. BMC Genom. 2018, 19, 622. [Google Scholar] [CrossRef] [Green Version]





| Protein Name | Gene ID | Reproductive Function | Source |
|---|---|---|---|
| Plasma membrane calcium-transporting ATPase | PMCA4 | Sperm hyperactivation and acrosome reaction | Al-Dossary, A.A. (2013) [25] |
| Pregnancy zone protein | PZP | Suppress T-cell function to prevent fetal rejection; Efficiently inhibits the aggregation of misfolded proteins during pregnancy | Skornicka, E. (2004) [40] Cater, H.J. (2019) [41] |
| Profilin 1 | PFN1 | Actin assembly regulator, essential for cell division and survival during embryogenesis | Witke, W. (2001) [42] |
| Disintegrin and metalloproteinase domain-containing protein | ADAM10 | Regulate Notch signaling, early embryonic cardiovascular development | Zhang, C. (2010) [43] |
| Gelsolin | GSN | Actin-severing protein regulates acrosome reaction and sperm capacitation | Finkelstein, M. (2010) [44] |
| Tetraspanin CD9 | CD9 | Egg membrane protein essential for sperm-egg fusion | Miyado, K. (2000) [45] |
| Heat shock 70 kDa protein 2 | HSPA2 | Sperm fertilization ability | Feng, H.L (2001) [46] |
| Heat shock 90 kDa protein | HSP90 | Modulate sperm capacitation via Erk1/2 and p38 MAPK signaling | Sun, P. (2021) [47] |
| Tubulin beta-4B chain | TUBB4B | Ciliary motility; egg, gamete transport | Yuan, S. (2021) [48] |
| Serpin Family G Member 1 | SERPING1 | Endometrial receptivity, implantation | Mirkin, S. (2005) [49] |
| Annexin A2 | ANXA2 | Critical for embryo adhesiveness to the human endometrium by RhoA activation | Garrido-Gómez, T. (2012) [50] |
| Protein Name | Gene ID | Function | Source |
|---|---|---|---|
| CD44 antigen | CD44 | Induce cell proliferation, increase cell survival and cellular motility | Senbanjo, L. (2017) [51] |
| Ras-related nuclear protein | RAN | Promote membrane targeting and stabilization of RhoA to orchestrate ovarian cancer cell invasion | Zaoui, K. (2019) [52] |
| Integrin beta | ITGB1 | Promote ovarian tumor progression and metastasis | Akinjiyan, F. (2022) [53] |
| S100 calcium binding protein A6 | S100A6 | Predict peritoneal tumor burden and is associated with advanced stage in ovarian cancer | Wei B. (2009) [54] |
| Versican core protein | VCAN | Modulate cell adhesion, proliferation, apoptosis, angiogenesis, invasion, and metastasis | Russo, A. (2022) [27] |
| Periostin | POSTN | Ovarian cancer migration and adhesion, wound healing | Yue, H (2021) [55] |
| The Ras homologous (Rho) protein | RHOs | Promote ovarian cancer progression and chemoresistance | Jeong, K.J. (2012) [56] Sharma, S. (2014) [57] |
| Transforming growth factor, beta-induced | TGFBI | Promote ovarian cancer migration and contributes to an Immunosuppressive Microenvironment | Lecker S.M.L. (2021) [58] |
| Aldehyde dehydrogenase 1A1 | ALDH1A1 | ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling | Meng, E. (2014) [59] |
| Lysosome-associated membrane glycoprotein 2 | LAMP2 | Regulate lysosomal stability as well as in autophagy | Eskelinen, E. (2002) [60] |
| Ras-related protein Rap-1b | RAP1B | The member of RAS oncogene family, promotes ovarian cancer metastasis via notch signaling | Lu, L (2016) [61] |
| Ras-related protein Rap-1A | RAP1A | ||
| Fibronectin | FN1 | Promote ovarian cancer invasion and metastasis through an α5β1-integrin/c-Met/FAK/Src-dependent signaling pathway, transducing signals through c-Met in an HGF/SF-independent manner | Lengyel, E (2010) [62] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zha, D.; Rayamajhi, S.; Sipes, J.; Russo, A.; Pathak, H.B.; Li, K.; Sardiu, M.E.; Bantis, L.E.; Mitra, A.; Puri, R.V.; et al. Proteomic Profiling of Fallopian Tube-Derived Extracellular Vesicles Using a Microfluidic Tissue-on-Chip System. Bioengineering 2023, 10, 423. https://doi.org/10.3390/bioengineering10040423
Zha D, Rayamajhi S, Sipes J, Russo A, Pathak HB, Li K, Sardiu ME, Bantis LE, Mitra A, Puri RV, et al. Proteomic Profiling of Fallopian Tube-Derived Extracellular Vesicles Using a Microfluidic Tissue-on-Chip System. Bioengineering. 2023; 10(4):423. https://doi.org/10.3390/bioengineering10040423
Chicago/Turabian StyleZha, Didi, Sagar Rayamajhi, Jared Sipes, Angela Russo, Harsh B. Pathak, Kailiang Li, Mihaela E. Sardiu, Leonidas E. Bantis, Amrita Mitra, Rajni V. Puri, and et al. 2023. "Proteomic Profiling of Fallopian Tube-Derived Extracellular Vesicles Using a Microfluidic Tissue-on-Chip System" Bioengineering 10, no. 4: 423. https://doi.org/10.3390/bioengineering10040423
APA StyleZha, D., Rayamajhi, S., Sipes, J., Russo, A., Pathak, H. B., Li, K., Sardiu, M. E., Bantis, L. E., Mitra, A., Puri, R. V., Trinidad, C. V., Cain, B. P., Isenberg, B. C., Coppeta, J., MacLaughlan, S., Godwin, A. K., & Burdette, J. E. (2023). Proteomic Profiling of Fallopian Tube-Derived Extracellular Vesicles Using a Microfluidic Tissue-on-Chip System. Bioengineering, 10(4), 423. https://doi.org/10.3390/bioengineering10040423