Delivery of dCas9 Activator System Using Magnetic Nanoparticles Technology as a Vector Delivery Method for Human Skin Fibroblast
Abstract
:1. Introduction
2. Materials and Methods
2.1. Guide RNA Design and dCas9 Activator Plasmid
2.2. Synthesis of SPIONs
2.3. Synthesis of PEI-Coated MNPs Complexed with CRISPR-Cas9 Plasmid
2.4. Physicochemical Characterization
2.5. Isolation and Primary Culture of Dermal Fibroblasts
2.6. Cell Culture
2.7. Magnetofection
2.8. Evaluation of Transfection Efficiency by Fluorescent Microscopy
2.9. Flow Cytometer Analyses
2.10. Real-Time RT-PCR (qRT-PCR)
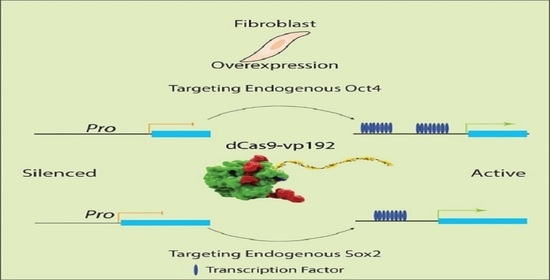
2.11. CRISPR-Mediated Activation of OCT4 and SOX2 Promoters in Human Foreskin Fibroblast
2.12. Toxicity Assay
2.13. Statistical Analysis
3. Results
3.1. Synthesis of CRISPR/Cas9-PEI-MNPs Complex
3.2. Analysis of Physicochemical Properties and Stability of CRISPR/Cas9-PEI-MNPs’ Complex
3.3. Scanning Electron Microscopy
3.4. Result of HFF Isolation
3.5. Effect of Magnetic Field on Cellular Uptake and GFP Expression
3.6. Cytotoxicity
3.7. Activation of Endogenous OCT4 and SOX2
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; White, R.R.; Fischer, K.E.; Zhang, Z.; Austad, S.N.; Vijg, J. Inducible aging in Hydra oligactis implicates sexual reproduction, loss of stem cells, and genome maintenance as major pathways. GeroScience 2020, 42, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Soufi, A.; Donahue, G.; Zaret, K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Rohiwal, S.; Dvorakova, N.; Klima, J.; Vaskovicova, M.; Senigl, F.; Slouf, M.; Pavlova, E.; Stepanek, P.; Babuka, D.; Benes, H. Polyethylenimine based magnetic nanoparticles mediated non-viral CRISPR/Cas9 system for genome editing. Sci. Rep. 2020, 10, 4619. [Google Scholar] [CrossRef] [PubMed]
- Balboa, D.; Weltner, J.; Eurola, S.; Trokovic, R.; Wartiovaara, K.; Otonkoski, T. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Rep. 2015, 5, 448–459. [Google Scholar] [CrossRef]
- Jinek, M. A Programmable Dual-Siswanto, FM, BH Kartiko/Media Sains 1 (2)(2017) 56 J. Media Sains–September 2017 RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Weltner, J.; Balboa, D.; Katayama, S.; Bespalov, M.; Krjutškov, K.; Jouhilahti, E.-M.; Trokovic, R.; Kere, J.; Otonkoski, T. Human pluripotent reprogramming with CRISPR activators. Nat. Commun. 2018, 9, 2643. [Google Scholar] [CrossRef]
- Hryhorowicz, M.; Grześkowiak, B.; Mazurkiewicz, N.; Śledziński, P.; Lipiński, D.; Słomski, R. Improved delivery of CRISPR/Cas9 system using magnetic nanoparticles into porcine fibroblast. Mol. Biotechnol. 2019, 61, 173–180. [Google Scholar] [CrossRef]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018, 36, 173–185. [Google Scholar] [CrossRef]
- Rajendrakumar, S.K.; Uthaman, S.; Cho, C.S.; Park, I.-K. Trigger-responsive gene transporters for anticancer therapy. Nanomaterials 2017, 7, 120. [Google Scholar] [CrossRef]
- Prijic, S.; Sersa, G. Magnetic nanoparticles as targeted delivery systems in oncology. Radiol. Oncol. 2011, 45, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-J.; Ke, J.-H.; Chen, G.-J.; Wang, L.-F. One-pot synthesis of PDMAEMA-bound iron oxide nanoparticles for magnetofection. J. Mater. Chem. B 2013, 1, 5916–5924. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, T.; Chen, H.; Liao, J.; Li, H.; Luo, Q.; Song, H.; Xiang, F.; Tan, J.; Zhou, J. Management strategies for the burn ward during COVID-19 pandemic. Burns 2020, 46, 756–761. [Google Scholar] [CrossRef]
- Bi, Q.; Song, X.; Hu, A.; Luo, T.; Jin, R.; Ai, H.; Nie, Y. Magnetofection: Magic magnetic nanoparticles for efficient gene delivery. Chin. Chem. Lett. 2020, 31, 3041–3046. [Google Scholar] [CrossRef]
- Lo, Y.-L.; Chou, H.-L.; Liao, Z.-X.; Huang, S.-J.; Ke, J.-H.; Liu, Y.-S.; Chiu, C.-C.; Wang, L.-F. Chondroitin sulfate-polyethylenimine copolymer-coated superparamagnetic iron oxide nanoparticles as an efficient magneto-gene carrier for microRNA-encoding plasmid DNA delivery. Nanoscale 2015, 7, 8554–8565. [Google Scholar] [CrossRef]
- Grześkowiak, B.F.; Hryhorowicz, M.; Tuśnio, K.; Grzeszkowiak, M.; Załęski, K.; Lipiński, D.; Zeyland, J.; Mykhaylyk, O.; Słomski, R.; Jurga, S. Generation of transgenic porcine fibroblast cell lines using nanomagnetic gene delivery vectors. Mol. Biotechnol. 2016, 58, 351–361. [Google Scholar] [CrossRef]
- Kim, J.B.; Sebastiano, V.; Wu, G.; Araúzo-Bravo, M.J.; Sasse, P.; Gentile, L.; Ko, K.; Ruau, D.; Ehrich, M.; van den Boom, D.; et al. Oct4-induced pluripotency in adult neural stem cells. Cell 2009, 136, 411–419. [Google Scholar] [CrossRef]
- Tiwari, A.P.; Rohiwal, S.S.; Suryavanshi, M.V.; Ghosh, S.J.; Pawar, S.H. Detection of the genomic DNA of pathogenic α-proteobacterium Ochrobactrum anthropi via magnetic DNA enrichment using pH responsive BSA@ Fe3O4 nanoparticles prior to in-situ PCR and electrophoretic separation. Microchim. Acta 2016, 183, 675–681. [Google Scholar] [CrossRef]
- Wightman, L.; Kircheis, R.; Rössler, V.; Carotta, S.; Ruzicka, R.; Kursa, M.; Wagner, E. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J. Gene Med. Cross-Discip. J. Res. Sci. Gene Transf. Clin. Appl. 2001, 3, 362–372. [Google Scholar]
- Zhang, L.; Li, Y.; Jimmy, C.Y.; Chen, Y.Y.; Chan, K.M. Assembly of polyethylenimine-functionalized iron oxide nanoparticles as agents for DNA transfection with magnetofection technique. J. Mater. Chem. B 2014, 2, 7936–7944. [Google Scholar] [CrossRef]
- Freeman, E.C.; Weiland, L.M.; Meng, W.S. Modeling the proton sponge hypothesis: Examining proton sponge effectiveness for enhancing intracellular gene delivery through multiscale modeling. J. Biomater. Sci. Polym. Ed. 2013, 24, 398–416. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Z.; Wang, X.; Xia, W.; Gu, H. Insights into the mechanism of magnetofection using MNPs-PEI/pDNA/free PEI magnetofectins. Int. J. Pharm. 2011, 419, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Pichon, C.; Gonçalves, C.; Midoux, P. Histidine-rich peptides and polymers for nucleic acids delivery. Adv. Drug Deliv. Rev. 2001, 53, 75–94. [Google Scholar] [CrossRef] [PubMed]
- Ota, S.; Takahashi, Y.; Tomitaka, A.; Yamada, T.; Kami, D.; Watanabe, M.; Takemura, Y. Transfection efficiency influenced by aggregation of DNA/polyethylenimine max/magnetic nanoparticle complexes. J. Nanoparticle Res. 2013, 15, 1653. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, L.; Ma, Y.; Li, X.; Gu, H. Control of aggregate size of polyethyleneimine-coated magnetic nanoparticles for magnetofection. Nano Res. 2009, 2, 365–372. [Google Scholar] [CrossRef]
- Steitz, B.; Hofmann, H.; Kamau, S.W.; Hassa, P.O.; Hottiger, M.O.; von Rechenberg, B.; Hofmann-Amtenbrink, M.; Petri-Fink, A. Characterization of PEI-coated superparamagnetic iron oxide nanoparticles for transfection: Size distribution, colloidal properties and DNA interaction. J. Magn. Magn. Mater. 2007, 311, 300–305. [Google Scholar] [CrossRef]
- Li, L.; Hu, S.; Chen, X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. [Google Scholar] [CrossRef]
- Namgung, R.; Singha, K.; Yu, M.K.; Jon, S.; Kim, Y.S.; Ahn, Y.; Park, I.-K.; Kim, W.J. Hybrid superparamagnetic iron oxide nanoparticle-branched polyethylenimine magnetoplexes for gene transfection of vascular endothelial cells. Biomaterials 2010, 31, 4204–4213. [Google Scholar] [CrossRef]
- Tomitaka, A.; Koshi, T.; Hatsugai, S.; Yamada, T.; Takemura, Y. Magnetic characterization of surface-coated magnetic nanoparticles for biomedical application. J. Magn. Magn. Mater. 2011, 323, 1398–1403. [Google Scholar] [CrossRef]
- Bajaj, A.; Kondaiah, P.; Bhattacharya, S. Synthesis and gene transfection efficacies of PEI− cholesterol-based lipopolymers. Bioconjugate Chem. 2008, 19, 1640–1651. [Google Scholar] [CrossRef]
- Sadeghi, Z.; Maleki, P.; Shahabi, F.; Bondarkhilli, S.A.M.; Masoumi, M.; Taheri, M.; Mohammadi, M.; Raheb, J. Surface modification of superparamagnetic iron oxide (SPION) and comparison of cytotoxicity effect of mPEG2000-PEI-SPION and mPEG750-PEI-SPION on the human embryonic carcinoma stem cell, NTERA2 cell line. Hum. Antibodies 2020, 28, 159–167. [Google Scholar] [CrossRef]
- Arsianti, M.; Lim, M.; Marquis, C.P.; Amal, R. Assembly of polyethylenimine-based magnetic iron oxide vectors: Insights into gene delivery. Langmuir 2010, 26, 7314–7326. [Google Scholar] [CrossRef]
- Lee, M.H.; Lin, C.C.; Thomas, J.L.; Li, J.A.; Lin, H.Y. Cellular reprogramming with multigene activation by the delivery of CRISPR/dCas9 ribonucleoproteins via magnetic peptide-imprinted chitosan nanoparticles. Mater. Today Bio 2021, 9, 100091. [Google Scholar] [CrossRef]







| sgRNA Name | Target Sequence (5′-3′) |
|---|---|
| OCT4 (1) | GGGGGAGAAACTGAGGCGA |
| OCT4 (2) | GACACAACTGGCGCCCCTCC |
| SOX2 (1) | TCTGTGGGGGACCTGCACTG |
| SOX2 (2) | GGCACAGTGCCAGAGGTCTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadi Ghanbarlou, M.; Abdoli, S.; Omid, H.; Qazizadeh, L.; Bamehr, H.; Raigani, M.; Shahsavarani, H.; Karimipour, M.; Shokrgozar, M.A. Delivery of dCas9 Activator System Using Magnetic Nanoparticles Technology as a Vector Delivery Method for Human Skin Fibroblast. Magnetochemistry 2023, 9, 71. https://doi.org/10.3390/magnetochemistry9030071
Mohammadi Ghanbarlou M, Abdoli S, Omid H, Qazizadeh L, Bamehr H, Raigani M, Shahsavarani H, Karimipour M, Shokrgozar MA. Delivery of dCas9 Activator System Using Magnetic Nanoparticles Technology as a Vector Delivery Method for Human Skin Fibroblast. Magnetochemistry. 2023; 9(3):71. https://doi.org/10.3390/magnetochemistry9030071
Chicago/Turabian StyleMohammadi Ghanbarlou, Mahdi, Shahriyar Abdoli, Hamed Omid, Leila Qazizadeh, Hadi Bamehr, Mozhgan Raigani, Hosein Shahsavarani, Morteza Karimipour, and Mohammad Ali Shokrgozar. 2023. "Delivery of dCas9 Activator System Using Magnetic Nanoparticles Technology as a Vector Delivery Method for Human Skin Fibroblast" Magnetochemistry 9, no. 3: 71. https://doi.org/10.3390/magnetochemistry9030071
APA StyleMohammadi Ghanbarlou, M., Abdoli, S., Omid, H., Qazizadeh, L., Bamehr, H., Raigani, M., Shahsavarani, H., Karimipour, M., & Shokrgozar, M. A. (2023). Delivery of dCas9 Activator System Using Magnetic Nanoparticles Technology as a Vector Delivery Method for Human Skin Fibroblast. Magnetochemistry, 9(3), 71. https://doi.org/10.3390/magnetochemistry9030071