Convolutional Neural Network Classification of Exhaled Aerosol Images for Diagnosis of Obstructive Respiratory Diseases
Abstract
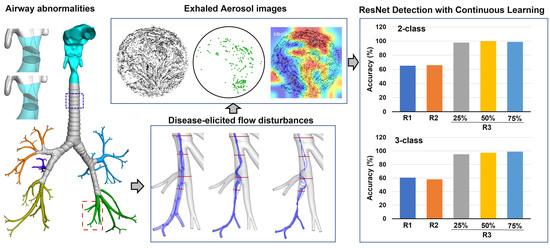
:1. Introduction
- (1)
- To assess model capacity in classifying images inside and outside the design space;
- (2)
- To quantify the benefits of continuous learning on the model’s performance;
- (3)
- To evaluate the relative importance of breath test variables on classification decisions;
- (4)
- To select an appropriate CNN model for the future development of obstructive lung diagnostic systems based on exhaled aerosol images.
2. Methods
2.1. Normal and Diseased Airway Models
2.2. Numerical Methods for Image Generation
2.3. Data Architecture
2.4. Design of CNN Model Training/Testing
3. Results
3.1. Exhaled Aerosol Images at Mouth Opening
3.1.1. Cumulative Aerosol Images
3.1.2. Disease-Associated Aerosol Distributions
3.2. Round 1 Training/Testing
3.2.1. Test Data with Decreasing Similarities
3.2.2. Comparison of Model Performance
3.3. Continous Training/Testing
3.3.1. Round 2
3.3.2. Round 3, 25% Outbox
3.3.3. Round 3, 50% Outbox
3.3.4. Outbox-Tested ROC Curves: Round 2 vs. 3
3.3.5. ResNet-50
3.4. Model Performance on New Test Datasets (Inbox_dp and Outbox_dp)
3.4.1. Inbox_dp vs. Outbox_Q_dp
3.4.2. Different Models on Outbox_Q_dp
3.4.3. ROC on Outbox_Q_dp
3.5. Heat Map and ReLU Features
4. Discussion
4.1. Model Sensitivity to Small Airway Remodeling
4.2. Geometrical, Breathing, and Aerosol Effects on Classification Decision
4.3. Model Evaluation and Continous Learning
4.4. Limitations
5. Conclusions
- (1)
- All models showed reasonably high classification accuracy on inbox images; the accuracy decreased notably on outbox images, with the magnitude varying with models;
- (2)
- ResNet-50 was the most robust among the four models when tested on both inbox and outbox images and for both diagnostic (2-class: normal vs. disease) and staging (3-class: D0, D1, D2) purposes;
- (3)
- CNN models could detect small airway remodeling (<1 mm) amidst a variety of variants (including glottal aperture changes of larger magnitudes, i.e., 3 mm);
- (4)
- Variation in flow rate was observed to be more important than throat opening and particle size in classification decisions;
- (5)
- Continuous learning significantly improved classification accuracy, with the relevance of training data strongly correlating with model performance.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darquenne, C. Aerosol deposition in health and disease. J. Aerosol Med. Pulm. Drug Deliv. 2012, 25, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Xi, J.; Kim, J.; Si, X.A.; Corley, R.A.; Kabilan, S.; Wang, S. CFD modeling and image analysis of exhaled aerosols due to a growing bronchial tumor: Towards non-invasive diagnosis and treatment of respiratory obstructive diseases. Theranostics 2015, 5, 443–455. [Google Scholar] [CrossRef]
- Lee, D.Y.; Lee, J.W. Dispersion during exhalation of an aerosol bolus in a double bifurcation. J. Aerosol Sci. 2001, 32, 805–815. [Google Scholar] [CrossRef]
- Darquenne, C.; Prisk, G.K. The effect of aging on aerosol bolus deposition in the healthy adult lung: A 19-year longitudinal study. J. Aerosol Med. Pulm. Drug Deliv. 2020, 33, 133–139. [Google Scholar] [CrossRef]
- Schwarz, K.; Biller, H.; Windt, H.; Koch, W.; Hohlfeld, J.M. Characterization of exhaled particles from the human lungs in airway obstruction. J. Aerosol Med. Pulm. Drug Deliv. 2015, 28, 52–58. [Google Scholar] [CrossRef]
- Kohlhäufl, M.; Brand, P.; Rock, C.; Radons, T.; Scheuch, G.; Meyer, T.; Schulz, H.; Pfeifer, K.J.; Häussinger, K.; Heyder, J. Noninvasive diagnosis of emphysema. Aerosol morphometry and aerosol bolus dispersion in comparison to HRCT. Am. J. Respir. Crit. Care Med. 1999, 160, 913–918. [Google Scholar] [CrossRef]
- Verbanck, S.; Schuermans, D.; Paiva, M.; Vincken, W. Saline aerosol bolus dispersion. II. The effect of conductive airway alteration. J. Appl. Physiol. 2001, 90, 1763–1769. [Google Scholar] [CrossRef] [Green Version]
- Sturm, R. Aerosol bolus dispersion in healthy and asthmatic children-theoretical and experimental results. Ann. Transl. Med. 2014, 2, 47. [Google Scholar]
- Xi, J.; Kim, J.; Si, X.A.; Zhou, Y. Diagnosing obstructive respiratory diseases using exhaled aerosol fingerprints: A feasibility study. J. Aerosol Sci. 2013, 64, 24–36. [Google Scholar] [CrossRef]
- Anderson, P.J.; Dolovich, M.B. Aerosols as diagnostic tools. J. Aerosol Med. 1994, 7, 77–88. [Google Scholar] [CrossRef]
- Blanchard, J.D. Aerosol bolus dispersion and aerosol-derived airway morphometry: Assessment of lung pathology and response to therapy, Part 1. J. Aerosol Med. 1996, 9, 183–205. [Google Scholar] [CrossRef]
- Löndahl, J.; Jakobsson, J.K.; Broday, D.M.; Aaltonen, H.L.; Wollmer, P. Do nanoparticles provide a new opportunity for diagnosis of distal airspace disease? Int. J. Nanomed. 2017, 12, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Inage, T.; Nakajima, T.; Yoshino, I.; Yasufuku, K. Early Lung Cancer Detection. Clin. Chest Med. 2018, 39, 45–55. [Google Scholar] [CrossRef]
- Roointan, A.; Ahmad Mir, T.; Ibrahim Wani, S.; Mati Ur, R.; Hussain, K.K.; Ahmed, B.; Abrahim, S.; Savardashtaki, A.; Gandomani, G.; Gandomani, M.; et al. Early detection of lung cancer biomarkers through biosensor technology: A review. J. Pharm. Biomed. Anal. 2019, 164, 93–103. [Google Scholar] [CrossRef]
- Blandin Knight, S.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef] [Green Version]
- Dama, E.; Colangelo, T.; Fina, E.; Cremonesi, M.; Kallikourdis, M.; Veronesi, G.; Bianchi, F. Biomarkers and lung cancer early detection: State of the art. Cancers 2021, 13, 3919. [Google Scholar] [CrossRef]
- Eggert, J.A.; Palavanzadeh, M.; Blanton, A. Screening and early detection of lung cancer. Semin. Oncol. Nurs. 2017, 33, 129–140. [Google Scholar] [CrossRef]
- Si, X.A.; Xi, J. Deciphering exhaled aerosol fingerprints for early diagnosis and personalized therapeutics of obstructive respiratory diseases in small airways. J. Nanotheranostics 2021, 2, 94–117. [Google Scholar] [CrossRef]
- Xi, J.; Zhao, W.; Yuan, J.E.; Kim, J.; Si, X.; Xu, X. Detecting lung diseases from exhaled aerosols: Non-invasive lung diagnosis using fractal analysis and SVM classification. PLoS ONE 2015, 10, e0139511. [Google Scholar] [CrossRef] [Green Version]
- Xi, J.; Zhao, W. Correlating exhaled aerosol images to small airway obstructive diseases: A study with dynamic mode decomposition and machine learning. PLoS ONE 2019, 14, e0211413. [Google Scholar] [CrossRef] [Green Version]
- Xi, J.; Si, X.A.; Kim, J.; Mckee, E.; Lin, E.-B. Exhaled aerosol pattern discloses lung structural abnormality: A sensitivity study using computational modeling and fractal analysis. PLoS ONE 2014, 9, e104682. [Google Scholar] [CrossRef] [Green Version]
- Valverde, J.M.; Imani, V.; Abdollahzadeh, A.; de Feo, R.; Prakash, M.; Ciszek, R.; Tohka, J. Transfer learning in magnetic resonance brain imaging: A systematic review. J. Imaging 2021, 7, 66. [Google Scholar] [CrossRef]
- Ayana, G.; Dese, K.; Choe, S.W. Transfer learning in breast cancer diagnoses via ultrasound imaging. Cancers 2021, 13, 738. [Google Scholar] [CrossRef]
- Gao, Y.; Cui, Y. Deep transfer learning for reducing health care disparities arising from biomedical data inequality. Nat. Commun. 2020, 11, 5131. [Google Scholar] [CrossRef]
- Link, J.; Perst, T.; Stoeve, M.; Eskofier, B.M. Wearable sensors for activity recognition in ultimate frisbee using convolutional neural networks and transfer learning. Sensors 2022, 22, 2560. [Google Scholar] [CrossRef]
- Maray, N.; Ngu, A.H.; Ni, J.; Debnath, M.; Wang, L. Transfer learning on small datasets for improved fall detection. Sensors 2023, 23, 1105. [Google Scholar] [CrossRef]
- Xi, J.; Zhao, W.; Yuan, J.E.; Cao, B.; Zhao, L. Multi-resolution classification of exhaled aerosol images to detect obstructive lung diseases in small airways. Comput. Biol. Med. 2017, 87, 57–69. [Google Scholar] [CrossRef]
- Xi, J.; Wang, Z.; Talaat, K.; Glide-Hurst, C.; Dong, H.J.S. Numerical study of dynamic glottis and tidal breathing on respiratory sounds in a human upper airway model. Sleep Breath. 2018, 22, 463–479. [Google Scholar] [CrossRef] [Green Version]
- U.S. Preventive Services Task Force. Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 325, 962–970. [Google Scholar] [CrossRef]
- Talaat, M.; Si, X.A.; Dong, H.; Xi, J. Leveraging statistical shape modeling in computational respiratory dynamics: Nanomedicine delivery in remodeled airways. Comput. Methods Programs Biomed. 2021, 204, 106079. [Google Scholar] [CrossRef]
- Xi, J.; Talaat, M.; Si, X.A.; Chandra, S. The application of statistical shape modeling for lung morphology in aerosol inhalation dosimetry. J. Aerosol Sci. 2021, 151, 105623. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Girshick, R.; Dollar, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5987–5995. [Google Scholar]
- Loey, M.; Manogaran, G.; Taha, M.H.N.; Khalifa, N.E.M. Fighting against COVID-19: A novel deep learning model based on YOLO-v2 with ResNet-50 for medical face mask detection. Sustain. Cities Soc. 2021, 65, 102600. [Google Scholar] [CrossRef]
- Benali Amjoud, A.; Amrouch, M. Convolutional neural networks backbones for object detection. In Image and Signal Processing, Proceedings of the 9th International Conference, ICISP 2020, Marrakesh, Morocco, 4–6 June 2020; Springer: Berlin/Heidelberg, Germany, 2020; Volume 12119. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. arXiv 2021, arXiv:2104.00298v00293. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Bansal, N.; Aljrees, T.; Yadav, D.P.; Singh, K.U.; Kumar, A.; Verma, G.K.; Singh, T. Real-time advanced computational intelligence for deep fake video detection. Appl. Sci. 2023, 13, 3095. [Google Scholar] [CrossRef]
- Wang, Z.; Shi, Z.; Tong, J.; Gong, W.; Wu, Z. A detection method for impact point water columns based on improved YOLO X. AIP Adv. 2022, 12, 065011. [Google Scholar] [CrossRef]
- Michele, A.; Colin, V.; Santika, D.D. MobileNet convolutional neural networks and support vector machines for palmprint recognition. Procedia Comput. Sci. 2019, 157, 110–117. [Google Scholar] [CrossRef]
- Xiao, Q.; Stewart, N.J.; Willmering, M.M.; Gunatilaka, C.C.; Thomen, R.P.; Schuh, A.; Krishnamoorthy, G.; Wang, H.; Amin, R.S.; Dumoulin, C.L.; et al. Human upper-airway respiratory airflow: In vivo comparison of computational fluid dynamics simulations and hyperpolarized 129Xe phase contrast MRI velocimetry. PLoS ONE 2021, 16, e0256460. [Google Scholar] [CrossRef]
- Xi, J.; Kim, J.; Si, X.A.; Corley, R.A.; Zhou, Y. Modeling of inertial deposition in scaled models of rat and human nasal airways: Towards in vitro regional dosimetry in small animals. J. Aerosol Sci. 2016, 99, 78–93. [Google Scholar] [CrossRef]
- Si, X.; Talaat, M.; Xi, J. SARS CoV-2 virus-laden droplets coughed from deep lungs: Numerical quantification in a single-path whole respiratory tract geometry. Phys. Fluids 2021, 33, 023306. [Google Scholar]
- Talaat, M.; Si, X.; Tanbour, H.; Xi, J. Numerical studies of nanoparticle transport and deposition in terminal alveolar models with varying complexities. Med. One 2019, 4, e190018. [Google Scholar]
- Xi, J.; Talaat, M.J.N. Nanoparticle deposition in rhythmically moving acinar models with interalveolar septal apertures. J. Nanomater. 2019, 9, 1126. [Google Scholar] [CrossRef] [Green Version]
- Xi, J.; Si, X.A.; Dong, H.; Zhong, H. Effects of glottis motion on airflow and energy expenditure in a human upper airway model. Eur. J. Mech. B 2018, 72, 23–37. [Google Scholar] [CrossRef]
- Xi, J.; Walfield, B.; Si, X.A.; Bankier, A.A. Lung physiological variations in COVID-19 patients and inhalation therapy development for remodeled lungs. SciMedicine J. 2021, 3, 198–208. [Google Scholar] [CrossRef]
- Brand, P.; Rieger, C.; Schulz, H.; Beinert, T.; Heyder, J. Aerosol bolus dispersion in healthy subjects. Eur. Respir. J. 1997, 10, 460–467. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Darquenne, C. Aerosol bolus dispersion in acinar airways—Influence of gravity and airway asymmetry. J. Appl. Physiol. 2012, 113, 442–450. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.W.; Lee, D.Y.; Kim, W.S. Dispersion of an aerosol bolus in a double bifurcation. J. Aerosol Sci. 2000, 31, 491–505. [Google Scholar] [CrossRef]
- Wang, J.; Xi, J.; Han, P.; Wongwiset, N.; Pontius, J.; Dong, H. Computational analysis of a flapping uvula on aerodynamics and pharyngeal wall collapsibility in sleep apnea. J. Biomech. 2019, 94, 88–98. [Google Scholar] [CrossRef]











| Training | Testing | |||
|---|---|---|---|---|
| Level 1 | Level 2 | Level 3 | ||
| Round 1 | 90% Base | 10% Base | Inbox | Outbox |
| Round 2: (plus 90% Base) | Th1, Th2 | 10% Base | Inbox | Outbox |
| Round 3 (plus 90% Base, and Th1, Th2) | 25% Outbox | 10% Base | Inbox | Outbox |
| 50% Outbox | 10% Base | Inbox | Outbox | |
| 75% Outbox | 10% Base | Inbox | Outbox | |
| Round 1 | 2-Classes | 3-Classes | |||||
|---|---|---|---|---|---|---|---|
| Network | (%) | Level 1 | Inbox | Outbox | Level 1 | Inbox | Outbox |
| AlexNet | Accuracy | 100 | 98.88 | 58.49 | 99.24 | 83.52 | 47.07 |
| AUC | 100 | 99.89 | 63.86 | 100 | 100 | 59.63 | |
| Specificity | 100 | 99.17 | 60.61 | 98.90 | 76.11 | 32.83 | |
| Sensitivity | 100 | 98.28 | 55.16 | 100 | 98.85 | 69.44 | |
| Precision | 100 | 98.28 | 47.12 | 97.62 | 66.67 | 39.68 | |
| ResNet-50 | Accuracy | 100 | 99.63 | 65.12 | 99.24 | 82.77 | 60.65 |
| AUC | 100 | 100 | 75.10 | 100 | 99.98 | 84.06 | |
| Specificity | 100 | 100 | 73.74 | 98.90 | 74.44 | 43.69 | |
| Sensitivity | 100 | 98.85 | 51.59 | 100 | 100 | 87.30 | |
| Precision | 100 | 100 | 55.56 | 97.62 | 65.41 | 49.66 | |
| MobileNet | Accuracy | 99.24 | 96.6 3 | 60.19 | 97.73 | 73.40 | 45.8 |
| AUC | 99.76 | 99.68 | 67.58 | 100 | 99.02 | 70.53 | |
| Specificity | 100 | 99.44 | 51.26 | 96.70 | 65.28 | 26.26 | |
| Sensitivity | 97.56 | 90.80 | 74.21 | 100 | 90.23 | 76.59 | |
| Precision | 100 | 98.75 | 49.21 | 93.18 | 55.67 | 39.79 | |
| EfficientNet | Accuracy | 96.97 | 91.20 | 61.27 | 90.15 | 70.22 | 41.98 |
| AUC | 100 | 95.98 | 67.12 | 99.57 | 96.12 | 66.38 | |
| Specificity | 100 | 97.22 | 61.87 | 89.01 | 63.06 | 29.55 | |
| Sensitivity | 90.24 | 78.74 | 60.32 | 92.68 | 85.06 | 61.51 | |
| Precision | 100 | 93.20 | 50.17 | 79.17 | 52.67 | 35.71 | |
| Round 2 | 2-Classes | 3-Classes | |||||
|---|---|---|---|---|---|---|---|
| Network | (%) | Level 1 | Inbox | Outbox | Level 1 | Inbox | Outbox |
| AlexNet | Accuracy | 98.60 | 98.69 | 68.06 | 99.30 | 80.15 | 54.01 |
| AUC | 99.98 | 99.94 | 79.55 | 100 | 99.97 | 78.42 | |
| Specificity | 98.90 | 99.17 | 61.36 | 98.90 | 71.67 | 35.10 | |
| Sensitivity | 98.08 | 97.70 | 78.57 | 100 | 97.70 | 83.73 | |
| Precision | 98.08 | 98.27 | 56.41 | 98.11 | 62.50 | 45.09 | |
| ResNet-50 | Accuracy | 99.30 | 99.44 | 65.90 | 100 | 91.01 | 58.18 |
| AUC | 100 | 99.99 | 84.50 | 100 | 99.99 | 82.13 | |
| Specificity | 98.90 | 100 | 53.54 | 100 | 86.67 | 44.70 | |
| Sensitivity | 100 | 98.28 | 85.32 | 100 | 100 | 79.37 | |
| Precision | 98.11 | 100 | 53.88 | 100 | 78.38 | 47.73 | |
| MobileNet | Accuracy | 97.90 | 95.88 | 71.14 | 95.10 | 77.34 | 54.48 |
| AUC | 99.89 | 99.33 | 83.96 | 100 | 99.40 | 79.35 | |
| Specificity | 98.90 | 99.44 | 62.12 | 92.31 | 68.33 | 36.11 | |
| Sensitivity | 96.15 | 88.51 | 85.32 | 100 | 95.98 | 83.33 | |
| Precision | 98.04 | 98.72 | 58.90 | 88.14 | 59.43 | 45.36 | |
| EfficientNet | Accuracy | 97.90 | 93.07 | 61.88 | 93.0 | 68.73 | 56.33 |
| AUC | 99.87 | 98.19 | 71.12 | 99.81 | 97.06 | 77.21 | |
| Specificity | 97.80 | 96.94 | 58.08 | 91.21 | 61.39 | 48.48 | |
| Sensitivity | 98.81 | 85.06 | 67.86 | 96.15 | 83.91 | 68.65 | |
| Precision | 96.23 | 93.08 | 50.74 | 86.21 | 51.23 | 45.89 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Talaat, M.; Xi, J.; Tan, K.; Si, X.A.; Xi, J. Convolutional Neural Network Classification of Exhaled Aerosol Images for Diagnosis of Obstructive Respiratory Diseases. J. Nanotheranostics 2023, 4, 228-247. https://doi.org/10.3390/jnt4030011
Talaat M, Xi J, Tan K, Si XA, Xi J. Convolutional Neural Network Classification of Exhaled Aerosol Images for Diagnosis of Obstructive Respiratory Diseases. Journal of Nanotheranostics. 2023; 4(3):228-247. https://doi.org/10.3390/jnt4030011
Chicago/Turabian StyleTalaat, Mohamed, Jensen Xi, Kaiyuan Tan, Xiuhua April Si, and Jinxiang Xi. 2023. "Convolutional Neural Network Classification of Exhaled Aerosol Images for Diagnosis of Obstructive Respiratory Diseases" Journal of Nanotheranostics 4, no. 3: 228-247. https://doi.org/10.3390/jnt4030011
APA StyleTalaat, M., Xi, J., Tan, K., Si, X. A., & Xi, J. (2023). Convolutional Neural Network Classification of Exhaled Aerosol Images for Diagnosis of Obstructive Respiratory Diseases. Journal of Nanotheranostics, 4(3), 228-247. https://doi.org/10.3390/jnt4030011