LRRK2-Dependent Neurodegeneration in Parkinson’s Disease
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Cellular Aging".
Viewed by 44129Editors
Interests: movement disorders; Parkinson’s disease; levodopa-induced dyskinesia; basal ganglia; neurotransmitter release; LRRK2; autophagy; dopamine; glutamate; opioids; nociceptin/orphanin FQ; MAO-B inhibitors
Special Issues, Collections and Topics in MDPI journals
Interests: LRRK2; Parkinson’s disease; autophagy; synapse; alpha-synuclein
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
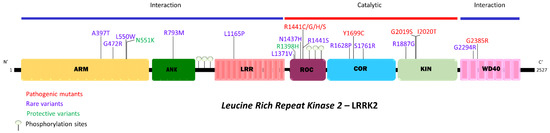
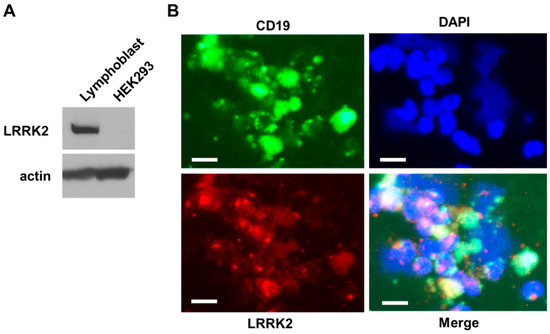
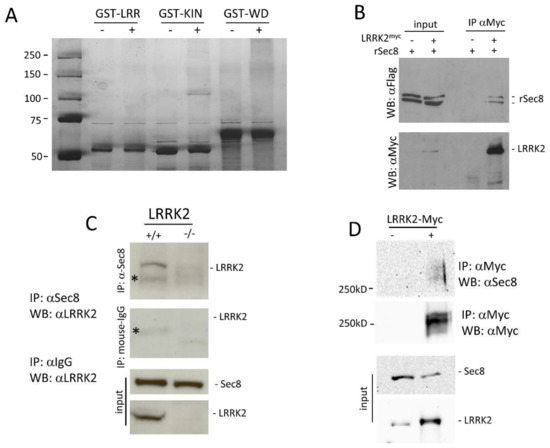
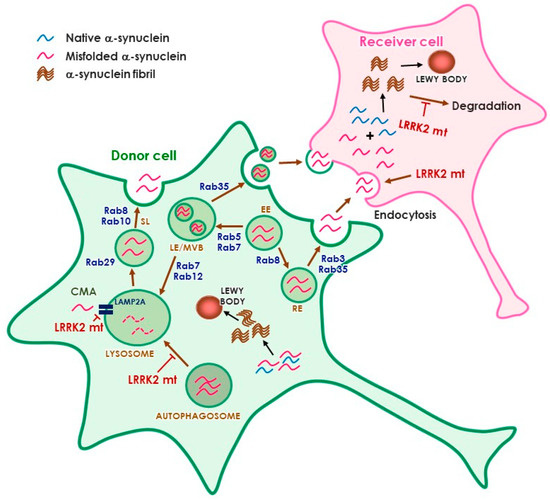
Parkinson’s disease is the second most common neurodegenerative disorder, affecting more than 10 million people worldwide. Parkinson’s disease has a complex etiology, with genetic factors playing a significant role not only in familiar, monogenic forms but also in idiopathic condition. Mutations in LRRK2 have been recognized as the most common genetic cause of familial Parkinson’s disease, and LRRK2 itself is considered a risk factor in idiopathic Parkinson’s disease. LRRK2 is a large multidomain protein with a GTPase and kinase catalytic core surrounded by protein–protein interaction domains. LRRK2 regulates several cellular functions, including vesicle trafficking, cytoskeletal dynamics, neurotransmitter release, synaptic plasticity, mitochondrial function, autophagy, and immune response. All of these functions are dysregulated in Parkinson’s disease, suggesting LRRK2 may play a direct or indirect role. Indeed, preclinical studies have revealed that pathogenic LRRK2 mutations, notably the p.G2019S substitution at the kinase domain, favor the degeneration of nigrostriatal dopaminergic neurons and formation of alpha-synuclein inclusions, which are neuropathological hallmarks of the disease. The enhancement of kinase activity proved to be instrumental for LRRK2-mediated neurodegeneration, leading to the development of LRRK2 kinase inhibitors as possible disease-modifying agents in Parkinson’s disease. In recognition of the intense and ever-growing research in the field, as well as the clinical relevance of this topic, this Topical Collection welcomes original papers and up-to-date reviews dealing with the mechanisms underlying the contribution of LRRK2 to the neurodegeneration associated with Parkinson’s disease.
Dr. Michele Morari
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- Parkinson’s disease
- LRRK2
- aging
- synucleinopathy
- proteostasis
- alpha-synuclein
- autophagy
- mitophagy
- microglia
- parkin
- PINK1
- neuroinflammation
- vesicle trafficking
- LRRK2 kinase inhibitors
- Rab