Ginkgolic Acid Rescues Lens Epithelial Cells from Injury Caused by Redox Regulated-Aberrant Sumoylation Signaling by Reviving Prdx6 and Sp1 Expression and Activities
Abstract
:1. Introduction
2. Results
2.1. GA, a Sumoylation Inhibitor, Augmented the Expression of Sp1 and Prdx6 in LECs
2.2. BA, a Sumoylation Agonist, Reduced the Expression of Sp1 and Prdx6
2.3. GA Inhibition of Global Protein Sumoylation Included Sp1 and Prdx6 Sumoylation in LECs In Vivo
2.4. BA Amplified Global Protein Sumoylation, Including Endogenous Sp1 and Prdx6 In Vivo
2.5. Oxidative Stress-Induced Aberrant Global Sumoylation of Proteins, Including Sp1 and Prdx6, Was Inhibited by GA in LECs In Vivo
2.6. GA Attenuated the Erratic Sumoylation Process in SRA-hLECs Overexpressing Sumo1 In Vivo
2.7. BA Further Promoted Protein Sumoylation in SRA-hLECs Overexpressing Sumo1 In Vivo
2.8. Treatment with Sumoylation Inhibitor GA Enhanced Sp1–DNA Binding, while Sumoylation Agonist BA Reduced the Binding
2.9. GA Significantly Promoted Sp1 Binding to Its Element by Blunting Oxidative Stress-Induced Aberrant Sumoylation Signaling
2.10. Prdx6 Transcription in LECs Was Significantly Potentiated by Sumoylation Inhibitor GA, and Was Inhibited by Sumoylation Agonist BA
2.11. GA Was Capable of Restoring Prdx6 Transcription Even in Cells Overexpressing Sumo1, while BA Further Reduced the Repression of Prdx6 Transcription
2.12. GA Delivery Blunted Oxidative Stress-Induced Deleterious Signaling-Mediated Injuries to LECs
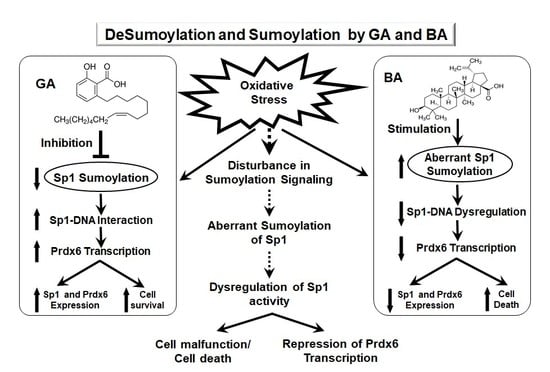
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Total Cell Extraction
4.3. Protein Expression Analysis
4.4. RT-qPCR
4.5. Sumoylation ELISA Assay
4.6. ChIP Assay
4.7. Plasmid and Constructs Detail
4.7.1. Construction of pEGFP-Sumo1
4.7.2. Site-Directed Mutagenesis (SDM)
4.8. CAT Reporter Assay
4.8.1. Construction of Human Prdx6 Promoter-CAT Reporter Vector
4.8.2. Cotransfection and Prdx6 Promoter Activity Assay
4.9. Quantitation of Intracellular ROS Level by H2-DCF-DA
4.10. Cell Survival Assay (MTS Assay)
4.11. Induction of Ultraviolet (UV) B and Hydrogen Peroxide (H2O2) Induced Stress
4.12. Statistical Methods
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Sp1 | Specificity protein 1 |
| Prdx6 | Peroxiredoxin 6 |
| GA | Ginkgolic acid |
| LECs | Lens epithelial cells |
| BA | Betulinic acid |
| SUMO | Small ubiquitin-related modifier |
| ROS | reactive oxygen species |
| Senps | Sumo-specific proteases |
| Prdxs | Peroxiredoxins |
| Cys | Cysteine |
| GSH | glutathione |
| PLA2 | phospholipase A2 |
| mLECs | mouse LECs |
| SRA-hLECs | human LEC cell line |
| h | hours |
| qPCR | quantitive polymerase chain reaction |
| RT-qPCR | real-time qeverse transcriptase-quantitive polymerase chain reaction |
| ELISA | enzyme-linked immunosorbent assay |
| DMSO | Dimethyl Sulfoxide |
| EGFP | enhanced green fluorescent protein |
| NEM | N-ethylmaleimide |
| HA | hemagglutinin |
| RIPA | radio immunoprecipitation assay |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| ChIP | chromatin immunoprecipitation |
| UV | Ultra violet |
| CAT | chloramphenicol acetyltransferase |
| DMEM | Dulbecco’s modified Eagle’s medium |
| MTS | 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2 to 4-sulphophenyl) 2H-tetrazolium salt |
| LEDGF | lens epithelial derived growth factor |
| AKI | acute kidney injury |
| CDKs | cyclin-dependent kinases |
| FBS | fetal bovine serum |
| min | minutes |
| s | seconds |
| DTT | 1,4-Dithiothreitol |
| CT | threshold cycle |
| OD | optical density |
| sec | seconds |
| SDM | Site-directed mutagenesis |
| H2DCFDA | dichlorofluorescin diacetate |
| PES | Phenazine ethosulfate |
References
- Johnson, E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004, 73, 355–382. [Google Scholar] [CrossRef] [PubMed]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, L.; Pakala, S.B.; Ohshiro, K.; Li, D.Q.; Kumar, R. SUMOylation and SUMO-interacting motif (SIM) of metastasis tumor antigen 1 (MTA1) synergistically regulate its transcriptional repressor function. J. Biol. Chem. 2011, 286, 43793–43808. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, I.; Ito, A.; Hirai, G.; Nishimura, S.; Kawasaki, H.; Saitoh, H.; Kimura, K.; Sodeoka, M.; Yoshida, M. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem. Biol. 2009, 16, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.Y.; Khachigian, L.M. Sp1 phosphorylation and its regulation of gene transcription. Mol. Cell. Biol. 2009, 29, 2483–2488. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.C.; Lee, C.C.; Yao, Y.L.; Lai, C.C.; Schmitz, M.L.; Yang, W.M. SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Sci. Rep. 2016, 6, 26509. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Wang, H.; Yang, X.X.; Geng, H.Y.; Gong, G.; Kim, H.J.; Zhou, Y.H.; Wu, J.J. Small Ubiquitin-Like Modifier 4 (SUMO4) Gene M55V Polymorphism and Type 2 Diabetes Mellitus: A Meta-analysis Including 6823 Subjects. Front. Endocrinol. 2017, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Kosoy, R.; Concannon, P. Functional variants in SUMO4, TAB2, and NFkappaB and the risk of type 1 diabetes. Genes Immun. 2005, 6, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Wang, Y.; Zhao, H.; Qin, L.; Shi, Y.; Zhu, X.; Song, L.; Zhou, X.; Chen, J.; Zhou, H.; et al. The critical role of SENP1-mediated GATA2 deSUMOylation in promoting endothelial activation in graft arteriosclerosis. Nat. Commun. 2017, 8, 15426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamoliatte, F.; McManus, F.P.; Maarifi, G.; Chelbi-Alix, M.K.; Thibault, P. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat. Commun. 2017, 8, 14109. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010, 428, 133–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhunchha, B.; Fatma, N.; Bhargavan, B.; Kubo, E.; Kumar, A.; Singh, D.P. Specificity protein, Sp1-mediated increased expression of Prdx6 as a curcumin-induced antioxidant defense in lens epithelial cells against oxidative stress. Cell Death Dis. 2011, 2, e234. [Google Scholar] [CrossRef] [PubMed]
- Chhunchha, B.; Fatma, N.; Kubo, E.; Singh, D.P. Aberrant sumoylation signaling evoked by reactive oxygen species impairs protective function of Prdx6 by destabilization and repression of its transcription. FEBS J. 2014, 281, 3357–3381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhunchha, B.; Kubo, E.; Fatma, N.; Singh, D.P. Sumoylation-deficient Prdx6 gains protective function by amplifying enzymatic activity and stability and escapes oxidative stress-induced aberrant Sumoylation. Cell Death Dis. 2017, 8, e2525. [Google Scholar] [CrossRef] [PubMed]
- Kubo, E.; Chhunchha, B.; Singh, P.; Sasaki, H.; Singh, D.P. Sulforaphane reactivates cellular antioxidant defense by inducing Nrf2/ARE/Prdx6 activity during aging and oxidative stress. Sci. Rep. 2017, 7, 14130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, E.; Jia, N.; Yan, M.; Ying, Z.; Sun, Q.; Wang, H.; Zhang, T.; Ma, X.; Ding, H.; Yao, X.; et al. SUMO-1 modification increases human SOD1 stability and aggregation. Biochem. Biophys. Res. Commun. 2006, 347, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Fatma, N.; Bhargavan, B.; Chhunchha, B.; Kubo, E.; Dey, S.; Takamura, Y.; Kumar, A.; Singh, D.P. Lens epithelium-derived growth factor deSumoylation by Sumo-specific protease-1 regulates its transcriptional activation of small heat shock protein and the cellular response. FEBS J. 2012, 279, 3048–3070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klenk, C.; Humrich, J.; Quitterer, U.; Lohse, M.J. SUMO-1 controls the protein stability and the biological function of phosducin. J. Biol. Chem. 2006, 281, 8357–8364. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.; Best, J.L.; Zon, L.I.; Gill, G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 2002, 10, 831–842. [Google Scholar] [CrossRef]
- Spengler, M.L.; Brattain, M.G. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J. Biol. Chem. 2006, 281, 5567–5574. [Google Scholar] [CrossRef] [PubMed]
- Zhong, N.; Xu, J. Synergistic activation of the human MnSOD promoter by DJ-1 and PGC-1alpha: regulation by SUMOylation and oxidation. Hum. Mol. Genet. 2008, 17, 3357–3367. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Wei, Q.; Su, Y.; Dong, Z. SUMOylation occurs in acute kidney injury and plays a cytoprotective role. Biochim. Biophys. Acta 2015, 1852, 482–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Lan, Y.; Xu, J.; Zhang, W.; Wen, Z. SUMO1-activating enzyme subunit 1 is essential for the survival of hematopoietic stem/progenitor cells in zebrafish. Development 2012, 139, 4321–4329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Yang, C.; Hong, Y.; Bi, H.; Zhao, F.; Liu, Y.; Ao, X.; Pang, P.; Xing, X.; Chang, A.K.; et al. The transcriptional activity of co-activator AIB1 is regulated by the SUMO E3 ligase PIAS1. Biol. Cell. 2012, 104, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, M.; Ao, X.; Chang, A.K.; Yang, C.; Zhao, F.; Bi, H.; Liu, Y.; Xiao, L.; Wu, H. CLOCK is a substrate of SUMO and sumoylation of CLOCK upregulates the transcriptional activity of estrogen receptor-alpha. Oncogene 2013, 32, 4883–4891. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J. Sumoylation regulates diverse biological processes. Cell. Mol. Life Sci. 2007, 64, 3017–3033. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Yu, T.; Huang, C.; Xia, X.; Liu, X.; Gu, J.; Xue, S.; Yeh, E.T.; Cheng, J. SUMO-specific protease 1 regulates mitochondrial biogenesis through PGC-1alpha. J. Biol. Chem. 2012, 287, 44464–44470. [Google Scholar] [CrossRef] [PubMed]
- Nayak, A.; Muller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 2014, 15, 422. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Bhargavan, B.; Chhunchha, B.; Kubo, E.; Kumar, A.; Fatma, N. Transcriptional protein Sp1 regulates LEDGF transcription by directly interacting with its cis-elements in GC-rich region of TATA-less gene promoter. PLoS ONE 2012, 7, e37012. [Google Scholar] [CrossRef] [PubMed]
- Peek, J.; Harvey, C.; Gray, D.; Rosenberg, D.; Kolla, L.; Levy-Myers, R.; Yin, R.; McMurry, J.L.; Kerscher, O. SUMO targeting of a stress-tolerant Ulp1 SUMO protease. PLoS ONE 2018, 13, e0191391. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, J.; Zuo, Y.; Deng, J.; Wang, L.S.; Chen, G.Q. SUMO-specific protease 1 regulates the in vitro and in vivo growth of colon cancer cells with the upregulated expression of CDK inhibitors. Cancer Lett. 2011, 309, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Ozkosem, B.; Feinstein, S.I.; Fisher, A.B.; O’Flaherty, C. Absence of Peroxiredoxin 6 Amplifies the Effect of Oxidant Stress on Mobility and SCSA/CMA3 Defined Chromatin Quality and Impairs Fertilizing Ability of Mouse Spermatozoa. Biol. Reprod. 2016, 94, 68. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.G.; Chae, H.Z.; Kim, K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 2005, 38, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. Peroxiredoxin 6: A bifunctional enzyme with glutathione peroxidase and phospholipase A(2) activities. Antioxid. Redox Signal. 2011, 15, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch. Biochem. Biophys. 2017, 617, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. The Phospholipase A2 Activity of Peroxiredoxin 6. J. Lipid Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Manevich, Y.; Fisher, A.B. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic. Biol. Med. 2005, 38, 1422–1432. [Google Scholar] [CrossRef] [PubMed]
- Chhunchha, B.; Fatma, N.; Kubo, E.; Rai, P.; Singh, S.P.; Singh, D.P. Curcumin abates hypoxia-induced oxidative stress based-ER stress-mediated cell death in mouse hippocampal cells (HT22) by controlling Prdx6 and NF-kappaB regulation. Am. J. Physiol. Cell Physiol. 2013, 304, C636–C655. [Google Scholar] [CrossRef] [PubMed]
- Stankovic-Valentin, N.; Drzewicka, K.; Konig, C.; Schiebel, E.; Melchior, F. Redox regulation of SUMO enzymes is required for ATM activity and survival in oxidative stress. EMBO J. 2016, 35, 1312–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhunchha, B.; Kubo, E.; Singh, P.; Singh, D.P. Sumoylation-deficient Prdx6 repairs aberrant Sumoylation-mediated Sp1 dysregulation-dependent Prdx6 repression and cell injury in aging and oxidative stress. Aging 2018, 10, 2284–2315. [Google Scholar] [CrossRef] [PubMed]
- Mango, D.; Weisz, F.; Nistico, R. Ginkgolic Acid Protects against Abeta-Induced Synaptic Dysfunction in the Hippocampus. Front. Pharmacol. 2016, 7, 401. [Google Scholar] [CrossRef] [PubMed]
- Chintharlapalli, S.; Papineni, S.; Ramaiah, S.K.; Safe, S. Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Res. 2007, 67, 2816–2823. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.I.; Wang, M.C.; Chen, S.Y.; Huang, S.T.; Yeh, Y.M.; Su, W.C.; Chang, W.C.; Hung, J.J. Betulinic acid decreases specificity protein 1 (Sp1) level via increasing the sumoylation of sp1 to inhibit lung cancer growth. Mol. Pharmacol. 2012, 82, 1115–1128. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Li, X.; Du, W.; Feng, Y.; Kong, X.; Li, Y.; Xiao, L.; Zhang, P. Antitumor effects of ginkgolic acid in human cancer cell occur via cell cycle arrest and decrease the Bcl-2/Bax ratio to induce apoptosis. Chemotherapy 2010, 56, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Zeng, Y.; Sun, R.R.; Liu, M.; Chen, S.; Zhang, P.Y. SUMOylation in cardiac disorders—A review. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1583–1587. [Google Scholar] [PubMed]
- Bossis, G.; Melchior, F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 2006, 21, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Yuan, H.; Liu, X.; Wang, H.; Chen, S.; Chen, Z.; de The, H.; Zhou, J.; Zhu, J. GATA5 SUMOylation is indispensable for zebrafish cardiac development. Biochim. Biophys. Acta 2017, 1861, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, J.; Lu, D.; Li, J.; Liu, M.; He, Y.; Williams, B.O.; Li, J.; Yang, T. Ginkgolic acid, a sumoylation inhibitor, promotes adipocyte commitment but suppresses adipocyte terminal differentiation of mouse bone marrow stromal cells. Sci. Rep. 2018, 8, 2545. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.T.; Craig, T.J.; Henley, J.M. SUMOylation of synapsin Ia maintains synaptic vesicle availability and is reduced in an autism mutation. Nat. Commun. 2015, 6, 7728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nistico, R.; Ferraina, C.; Marconi, V.; Blandini, F.; Negri, L.; Egebjerg, J.; Feligioni, M. Age-related changes of protein SUMOylation balance in the AbetaPP Tg2576 mouse model of Alzheimer’s disease. Front. Pharmacol. 2014, 5, 63. [Google Scholar] [PubMed]
- Zhou, Y.; Ji, C.; Cao, M.; Guo, M.; Huang, W.; Ni, W.; Meng, L.; Yang, H.; Wei, J.F. Inhibitors targeting the SUMOylation pathway: A patent review 20122015 (Review). Int. J. Mol. Med. 2018, 41, 3–12. [Google Scholar] [PubMed]
- Zhang, L.; Yang, T.H.; Li, D.W. Roles of SUMOylation in Heart Development and Cardiovascular Diseases. Curr. Mol. Med. 2017, 16, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, L.; Wen, S.; Zhu, H.; Yu, W.; Moskowitz, I.P.; Shaw, G.M.; Finnell, R.H.; Schwartz, R.J. Defective sumoylation pathway directs congenital heart disease. Birth Defects Res. A Clin. Mol. Teratol. 2011, 91, 468–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubey, A.; Marabotti, A.; Ramteke, P.W.; Facchiano, A. Interaction of human chymase with ginkgolides, terpene trilactones of Ginkgo biloba investigated by molecular docking simulations. Biochem. Biophys. Res. Commun. 2016, 473, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Marcelli, S.; Ficulle, E.; Piccolo, L.; Corbo, M.; Feligioni, M. An overview of the possible therapeutic role of SUMOylation in the treatment of Alzheimer’s disease. Pharmacol. Res. 2018, 130, 420–437. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Li, X.; Zhang, L.; Zhang, Y.; Lu, X.; Tian, J. Improved metabolites of pharmaceutical ingredient grade Ginkgo biloba and the correlated proteomics analysis. Proteomics 2015, 15, 1868–1883. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Zheng, J.; Jin, X.; Wei, G.; Wang, G.; Sun, X.; Li, X. Ginkgolic acid inhibits the invasiveness of colon cancer cells through AMPK activation. Oncol. Lett. 2017, 14, 5831–5838. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Duan, W.; Han, S.; Lei, J.; Xu, Q.; Chen, X.; Jiang, Z.; Nan, L.; Li, J.; Chen, K.; et al. Ginkgolic acid suppresses the development of pancreatic cancer by inhibiting pathways driving lipogenesis. Oncotarget 2015, 6, 20993–21003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamdoun, S.; Efferth, T. Ginkgolic acids inhibit migration in breast cancer cells by inhibition of NEMO sumoylation and NF-kappaB activity. Oncotarget 2017, 8, 35103–35115. [Google Scholar] [CrossRef] [PubMed]
- Ibaraki, N.; Chen, S.C.; Lin, L.R.; Okamoto, H.; Pipas, J.M.; Reddy, V.N. Human lens epithelial cell line. Exp. Eye Res. 1998, 67, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Wu, S.B.; Wu, Y.T.; Wei, Y.H. Oxidative stress response elicited by mitochondrial dysfunction: implication in the pathophysiology of aging. Exp. Biol. Med. 2013, 238, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.C.; Hung, J.J. Functional role of post-translational modifications of Sp1 in tumorigenesis. J. Biomed. Sci. 2012, 19, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, G. SUMO changes Sox for developmental diversity. Mol. Cell 2005, 20, 495–496. [Google Scholar] [CrossRef] [PubMed]
- Flotho, A.; Melchior, F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev. Biochem. 2013, 82, 357–385. [Google Scholar] [CrossRef] [PubMed]
- Srikumar, T.; Lewicki, M.C.; Costanzo, M.; Tkach, J.M.; van Bakel, H.; Tsui, K.; Johnson, E.S.; Brown, G.W.; Andrews, B.J.; Boone, C.; et al. Global analysis of SUMO chain function reveals multiple roles in chromatin regulation. J. Cell Biol. 2013, 201, 145–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewicki, M.C.; Srikumar, T.; Johnson, E.; Raught, B. The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. J. Proteomics 2015, 118, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Drisaldi, B.; Colnaghi, L.; Fioriti, L.; Rao, N.; Myers, C.; Snyder, A.M.; Metzger, D.J.; Tarasoff, J.; Konstantinov, E.; Fraser, P.E.; et al. SUMOylation Is an Inhibitory Constraint that Regulates the Prion-like Aggregation and Activity of CPEB3. Cell Rep. 2015, 11, 1694–1702. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Wilkinson, K.A.; Nishimune, A.; Henley, J.M. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2007, 8, 948–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarge, K.D.; Park-Sarge, O.K. SUMO and its role in human diseases. Int. Rev. Cell Mol. Biol. 2011, 288, 167–183. [Google Scholar] [PubMed]
- Sarge, K.D.; Park-Sarge, O.K. Sumoylation and human disease pathogenesis. Trends Biochem. Sci. 2009, 34, 200–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fatma, N.; Kubo, E.; Sharma, P.; Beier, D.R.; Singh, D.P. Impaired homeostasis and phenotypic abnormalities in Prdx6-/-mice lens epithelial cells by reactive oxygen species: increased expression and activation of TGFbeta. Cell Death Differ. 2005, 12, 734–750. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Du, X.; Meng, F.T.; Zhou, J.N. SUMO negatively regulates BACE expression. Neuro Endocrinol. Lett. 2011, 32, 313–316. [Google Scholar] [PubMed]
- Hua, Z.; Wu, C.; Fan, G.; Tang, Z.; Cao, F. The antibacterial activity and mechanism of ginkgolic acid C15:1. BMC Biotechnol. 2017, 17, 5. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Salzwedel, K.; Wang, D.; Chakravarty, S.; Freed, E.O.; Wild, C.T.; Li, F. A single polymorphism in HIV-1 subtype C SP1 is sufficient to confer natural resistance to the maturation inhibitor bevirimat. Antimicrob. Agents Chemother. 2011, 55, 3324–3329. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Lucas, B.; Molcho, E.; Schiff, T.; Vigodner, M. Inhibition of CDK1 activity by sumoylation. Biochem. Biophys. Res. Commun. 2016, 478, 919–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.P.; Kubo, E.; Takamura, Y.; Shinohara, T.; Kumar, A.; Chylack, L.T., Jr.; Fatma, N. DNA binding domains and nuclear localization signal of LEDGF: Contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J. Mol. Biol. 2006, 355, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Ohguro, N.; Chylack, L.T., Jr.; Shinohara, T. Lens epithelium-derived growth factor: Increased resistance to thermal and oxidative stresses. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1444–1451. [Google Scholar]
- Kubo, E.; Singh, D.P.; Fatma, N.; Akagi, Y. TAT-mediated peroxiredoxin 5 and 6 protein transduction protects against high-glucose-induced cytotoxicity in retinal pericytes. Life Sci. 2009, 84, 857–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, E.; Miyazawa, T.; Fatma, N.; Akagi, Y.; Singh, D.P. Development- and age-associated expression pattern of peroxiredoxin 6, and its regulation in murine ocular lens. Mech. Ageing Dev. 2006, 127, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Kubo, E.; Hasanova, N.; Tanaka, Y.; Fatma, N.; Takamura, Y.; Singh, D.P.; Akagi, Y. Protein expression profiling of lens epithelial cells from Prdx6-depleted mice and their vulnerability to UV radiation exposure. Am. J. Physiol. Cell Physiol. 2010, 298, C342–C354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doetzlhofer, A.; Rotheneder, H.; Lagger, G.; Koranda, M.; Kurtev, V.; Brosch, G.; Wintersberger, E.; Seiser, C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 1999, 19, 5504–5511. [Google Scholar] [CrossRef] [PubMed]
- Kubo, E.; Fatma, N.; Sharma, P.; Shinohara, T.; Chylack, L.T., Jr.; Akagi, Y.; Singh, D.P. Transactivation of involucrin, a marker of differentiation in keratinocytes, by lens epithelium-derived growth factor (LEDGF). J. Mol. Biol. 2002, 320, 1053–1063. [Google Scholar] [CrossRef]
- Cory, A.H.; Owen, T.C.; Barltrop, J.A.; Cory, J.G. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991, 3, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Kubo, E.; Fatma, N.; Akagi, Y.; Beier, D.R.; Singh, S.P.; Singh, D.P. TAT-mediated PRDX6 protein transduction protects against eye lens epithelial cell death and delays lens opacity. Am. J. Physiol. Cell Physiol. 2008, 294, C842–C855. [Google Scholar] [CrossRef] [PubMed]












© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chhunchha, B.; Singh, P.; Singh, D.P.; Kubo, E. Ginkgolic Acid Rescues Lens Epithelial Cells from Injury Caused by Redox Regulated-Aberrant Sumoylation Signaling by Reviving Prdx6 and Sp1 Expression and Activities. Int. J. Mol. Sci. 2018, 19, 3520. https://doi.org/10.3390/ijms19113520
Chhunchha B, Singh P, Singh DP, Kubo E. Ginkgolic Acid Rescues Lens Epithelial Cells from Injury Caused by Redox Regulated-Aberrant Sumoylation Signaling by Reviving Prdx6 and Sp1 Expression and Activities. International Journal of Molecular Sciences. 2018; 19(11):3520. https://doi.org/10.3390/ijms19113520
Chicago/Turabian StyleChhunchha, Bhavana, Prerna Singh, Dhirendra P. Singh, and Eri Kubo. 2018. "Ginkgolic Acid Rescues Lens Epithelial Cells from Injury Caused by Redox Regulated-Aberrant Sumoylation Signaling by Reviving Prdx6 and Sp1 Expression and Activities" International Journal of Molecular Sciences 19, no. 11: 3520. https://doi.org/10.3390/ijms19113520
APA StyleChhunchha, B., Singh, P., Singh, D. P., & Kubo, E. (2018). Ginkgolic Acid Rescues Lens Epithelial Cells from Injury Caused by Redox Regulated-Aberrant Sumoylation Signaling by Reviving Prdx6 and Sp1 Expression and Activities. International Journal of Molecular Sciences, 19(11), 3520. https://doi.org/10.3390/ijms19113520