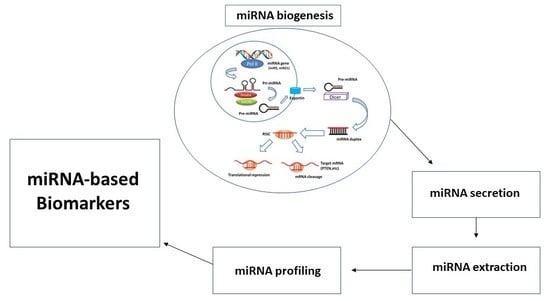
Insight toward the MicroRNA Profiling of Laryngeal Cancers: Biological Role and Clinical Impact
Abstract
:1. Introduction
2. miRNA Deregulation
2.1. Laryngeal Cancer
2.2. Other HNSCC
3. miRNAs as Biomarkers for the Prediction of Initiation and Progression
3.1. Laryngeal Cancer
3.2. Other HNSCC
4. miRNAs as Biomarkers for the Prognosis
4.1. Laryngeal Cancer
4.2. Other HNSCC
5. Resistance Related miRNAs
5.1. Radio-Resistance
5.2. Chemo-Resistance
5.3. Thermal-Resistance
6. Infection and Life Habit Related miRNAs
6.1. HPV Infection
6.2. Smoking Tobacco and Alcoholic Beverage
7. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| miRNA | MicroRNA |
| HNSCC | Head and neck squamous cell carcinoma |
| HNC | Head and neck cancer |
| LCa | Laryngeal cancer |
| qRT-PCR | Quantitative real-time PCR |
| RISC | RNA-induced silencing complex |
| OSCC | Oral squamous cell cancer |
| SACC | Salivary adenoid cystic carcinoma |
| MSSCC | Maxillary sinus squamous cell carcinoma |
| EMT | Epithelial-mesenchymal transition |
| OTSCC | Oral tongue squamous cell carcinoma |
| NPC | Nasopharyngeal carcinoma |
| TSCC | Tongue squamous cell carcinoma |
| SN-SCC | Sinonasal squamous cell carcinomas |
| HSCC | Hypopharyngeal squamous cell carcinoma |
| EBV | Epstein–Barr virus |
| LRC | Locoregional control |
| MDR | Multiple drug resistance |
| ACC | Adenoid cystic carcinoma |
| Pim-1 | Moloney murine leukemia virus 1 |
| AUC | Area under curve |
| PUMA | p53-upregulated modulator of apoptosis |
| ROC | Receiver operating characteristic |
| HPV | Human papillomavirus |
| SCC | Squamous cell carcinoma |
| TSCC/BOTSCC | Tonsillar and base of tongue cancer |
References
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Murray, T.; Thun, M.J. Cancer statistics, 2008. CA Cancer J. Clin. 2008, 58, 71–96. [Google Scholar] [CrossRef]
- Leemans, C.R.; Braakhuis, B.J.M.; Brakenhoff, R.H. Response to correspondence on the molecular biology of head and neck cancer. Nat. Rev. Cancer 2011, 11, 382. [Google Scholar] [CrossRef] [Green Version]
- Kulasinghe, A.; Perry, C.; Jovanović, L.; Nelson, C.; Punyadeera, C. Circulating tumour cells in metastatic head and neck cancers. Int. J. Cancer 2014, 136, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Babu, J.M.; Prathibha, R.; Jijith, V.; Hariharan, R.; Pillai, M.R. A miR-centric view of head and neck cancers. Biochim. Biophys. Acta Rev. Cancer 2011, 1816, 67–72. [Google Scholar] [CrossRef]
- Janiszewska, J.; Szaumkessel, M.; Szyfter-Harris, J. MicroRNAs are important players in head and neck carcinoma: A review. Crit. Rev. Oncol. 2013, 88, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Cabay, R.J.; Jin, Y.; Wang, A.; Lu, Y.; Shah-Khan, M.; Zhou, X. MicroRNA Deregulations in Head and Neck Squamous Cell Carcinomas. J. Oral Maxillofac. Res. 2013, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Bartel, B. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, H.; Zarone, M.R.; Lombardi, A.; Ricciardiello, F.; Caraglia, M.; Misso, G. Early detection of laryngeal cancer: Prominence of miRNA signature as a new tool for clinicians. Transl. Med. Rep. 2017, 1, 16502. [Google Scholar] [CrossRef] [Green Version]
- Misso, G.; Zarone, M.R.; Grimaldi, A.; di Martino, M.T.; Lombardi, A.; Kawasaki, H.; Stiuso, P.; Tassone, P.; Tagliaferri, P.; Caraglia, M. Non Coding RNAs: A New Avenue for the Self-Tailoring of Blood Cancer Treatment. Curr. Drug Targets 2016, 18, 35–55. [Google Scholar] [CrossRef] [PubMed]
- Syeda, Z.A.; Langden, S.; Munkhzul, C.; Lee, M.; Song, S. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.-H.; Lee, M.-J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zernecke, A.; Bidzhekov, K.; Noels, H.; Shagdarsuren, E.; Gan, L.; Denecke, B.; Hristov, M.; Köppel, T.; Nazari-Jahantigh, M.; Lutgens, E.; et al. Delivery of MicroRNA-126 by Apoptotic Bodies Induces CXCL12-Dependent Vascular Protection. Sci. Signal. 2009, 2, 81. [Google Scholar] [CrossRef]
- Arroyo, J.; Chevillet, J.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nature 2011, 13, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef]
- Guénel, P.; Chastang, J.F.; Luce, D.; Leclerc, A.; Brugere, J. A study of the interaction of alcohol drinking and tobacco smoking among French cases of laryngeal cancer. J. Epidemiol. Community Health 1988, 42, 350–354. [Google Scholar] [CrossRef] [Green Version]
- Falk, R.T.; Pickle, L.W.; Brown, L.M.; Mason, T.J.; A Buffler, P.; Fraumeni, J.F. Effect of smoking and alcohol consumption on laryngeal cancer risk in coastal Texas. Cancer Res. 1989, 49, 4024–4029. [Google Scholar]
- la Vecchia, C.; Negri, E.; D’Avanzo, B.; Franceschi, S.; de Carli, A.; Boyle, P. Dietary indicators of laryngeal cancer risk. Cancer Res. 1990, 50, 4497–4500. [Google Scholar]
- de Miguel-Luken, M.J.; Chaves-Conde, M.; Carnero, A. A genetic view of laryngeal cancer heterogeneity. Cell Cycle 2016, 15, 1202–1212. [Google Scholar] [CrossRef] [PubMed]
- Sessions, N.G. Surgical pathology of cancer of the larynx and hypopharynx. Laryngoscope 1976, 86, 814–839. [Google Scholar] [CrossRef] [PubMed]
- Lu, E.; Su, J.; Zeng, W.; Zhang, C. Enhanced miR-9 promotes laryngocarcinoma cell survival via down-regulating PTEN. Biomed. Pharmacother. 2016, 84, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, J.; Wang, B.; Ren, J.-C.; Su, W.; Zhang, T. MicroRNA-10b Triggers the Epithelial–Mesenchymal Transition (EMT) of Laryngeal Carcinoma Hep-2 Cells by Directly Targeting the E-cadherin. Appl. Biochem. Biotechnol. 2015, 176, 33–44. [Google Scholar] [CrossRef]
- Liu, M.; Wu, H.; Liu, T.; Li, Y.; Wang, F.; Wan, H.-Y.; Li, X.; Tang, H. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009, 19, 828–837. [Google Scholar] [CrossRef]
- Zheng, J.; Xue, H.; Wang, T.; Jiang, Y.; Liu, B.; Li, J.; Liu, Y.; Wang, W.; Zhang, B.; Sun, M. miR-21 downregulates the tumor suppressor P12CDK2AP1 and Stimulates Cell Proliferation and Invasion. J. Cell. Biochem. 2011, 112, 872–880. [Google Scholar] [CrossRef]
- Zhong, G.; Xiong, X. miR-205 promotes proliferation and invasion of laryngeal squamous cell carcinoma by suppressing CDK2AP1 expression. Boil. Res. 2015, 48, 60. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Ren, S.; Su, Z.; Liu, C.; Deng, T.; Huang, N.; Tian, Y.; Qiu, Y.; Liu, Y. Increased expression of miR-93 is associated with poor prognosis in head and neck squamous cell carcinoma. Tumor Boil. 2015, 36, 3949–3956. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Zhou, L.; Cao, P.; Gong, H.; Zhang, Y. MicroRNA-93 regulates cyclin G2 expression and plays an oncogenic role in laryngeal squamous cell carcinoma. Int. J. Oncol. 2014, 46, 161–174. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Wang, Z.; Song, Y.; Tai, X.-H.; Ji, W.-Y.; Gu, H. MicroRNA-126 modulates the tumor microenvironment by targeting calmodulin-regulated spectrin-associated protein 1 (Camsap1). Int. J. Oncol. 2014, 44, 1678–1684. [Google Scholar] [CrossRef] [Green Version]
- Lian, R.; Lu, B.; Jiao, L.; Li, S.; Wang, H.; Miao, W.; Yu, W. MiR-132 plays an oncogenic role in laryngeal squamous cell carcinoma by targeting FOXO1 and activating the PI3K/AKT pathway. Eur. J. Pharmacol. 2016, 792, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, B.; Zhao, X.-D.; Wang, L.-Y.; Ji, W. MicroRNA-221 accelerates the proliferation of laryngeal cancer cell line Hep-2 by suppressing Apaf-1. Oncol. Rep. 2015, 33, 1221–1226. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Gao, W.; Zhang, C.; Wen, S.; Huangfu, H.; Kang, J.; Wang, B. Hsa-miR-301a-3p Acts as an Oncogene in Laryngeal Squamous Cell Carcinoma via Target Regulation of Smad4. J. Cancer 2015, 6, 1260–1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, G.; Zhang, D.; Zheng, Y.; Wen, L.; Yu, D.; Lu, Y.; Zhao, Y. microRNA-423-3p promotes tumor progression via modulation of AdipoR2 in laryngeal carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 5683–5691. [Google Scholar]
- Wang, F.; Song, G.; Liu, M.; Li, X.; Tang, H. miRNA-1 targets fibronectin1 and suppresses the migration and invasion of the HEp2 laryngeal squamous carcinoma cell line. FEBS Lett. 2011, 585, 3263–3269. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Chen, Z.; Xue, F.; Chen, W.; Ma, R.; Cheng, S.; Cui, P. MicroRNA-24 inhibits growth, induces apoptosis, and reverses radioresistance in laryngeal squamous cell carcinoma by targeting X-linked inhibitor of apoptosis protein. Cancer Cell Int. 2015, 15, 61. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Li, L.; Feng, P.; Wan, J.; Li, J. Downregulation of miR-34a contributes to the proliferation and migration of laryngeal carcinoma cells by targeting cyclin D1. Oncol. Rep. 2016, 36, 390–398. [Google Scholar] [CrossRef]
- Maroof, H.; Salajegheh, A.; Smith, R.A.; Lam, A.K.-Y. MicroRNA-34 family, mechanisms of action in cancer: a review. Curr. Cancer Drug Targets 2014, 14, 737–751. [Google Scholar] [CrossRef] [Green Version]
- Cai, K.-M.; Bao, X.-L.; Kong, X.-H.; Jinag, W.; Mao, M.-R.; Chu, J.-S.; Huang, Y.-J.; Zhao, X.-J. Hsa-miR-34c suppresses growth and invasion of human laryngeal carcinoma cells via targeting c-Met. Int. J. Mol. Med. 2010, 25, 565–571. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ma, H.; Sun, J. microRNA-34a/c function as tumor suppressors in Hep-2 laryngeal carcinoma cells and may reduce GALNT7 expression. Mol. Med. Rep. 2014, 9, 1293–1298. [Google Scholar] [CrossRef]
- Wu, X.; Cui, C.-L.; Chen, W.-L.; Fu, Z.-Y.; Cui, X.-Y.; Gong, X. MiR-144 suppresses the growth and metastasis of laryngeal squamous cell carcinoma by targeting IRS1. Am. J. Transl. Res. 2016, 8, 1–11. [Google Scholar] [PubMed]
- Yu, W.; Zhang, G.; Lu, B.; Li, J.; Wu, Z.; Ma, H.; Wang, H.; Lian, R. MiR-340 impedes the progression of laryngeal squamous cell carcinoma by targeting EZH2. Gene 2016, 577, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Song, F.-C.; Yang, Y.; Liu, J.-X. Expression, and significances of MiRNA Let-7 and HMGA2 in laryngeal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4452–4458. [Google Scholar] [PubMed]
- Lu, Y.-C.; Chen, Y.-J.; Wang, H.-M.; Tsai, C.-Y.; Chen, W.-H.; Huang, Y.-C.; Fan, K.-H.; Tsai, C.-N.; Huang, S.-F.; Kang, C.-J.; et al. Oncogenic Function and Early Detection Potential of miRNA-10b in Oral Cancer as Identified by microRNA Profiling. Cancer Prev. Res. 2012, 5, 665–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Severino, P.; Brüggemann, H.; Andreghetto, F.M.; Camps, C.; Klingbeil, M.D.F.G.; Pereira, W.D.O.; Soares, R.M.; Moyses, R.; Filho, V.W.; Mathor, M.B.; et al. MicroRNA expression profile in head and neck cancer: HOX-cluster embedded microRNA-196a and microRNA-10b dysregulation implicated in cell proliferation. BMC Cancer 2013, 13, 533. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wu, Z.; Peng, Y.; Liu, X.; Lu, J.; Wang, L.; Pan, Q.; He, M.-L.; Li, X.-P. MicroRNA-10b induced by Epstein-Barr virus-encoded latent membrane protein-1 promotes the metastasis of human nasopharyngeal carcinoma cells. Cancer Lett. 2010, 299, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Rather, M.I.; Nagashri, M.N.; Swamy, S.S.; Gopinath, K.S.; Kumar, A. Oncogenic microRNA-155 down-regulates tumor suppressor CDC73 and promotes oral squamous cell carcinoma cell proliferation: Implications for cancer therapeutics. J. Biol. Chem. 2013, 288, 608–618. [Google Scholar] [CrossRef] [Green Version]
- Ni, Y.; Huang, X.-F.; Wang, Z.-Y.; Han, W.; Deng, R.-Z.; Mou, Y.-B.; Ding, L.; Hou, Y.; Hu, Q.-G. Upregulation of a potential prognostic biomarker, miR-155, enhances cell proliferation in patients with oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 117, 227–233. [Google Scholar] [CrossRef]
- Baba, O.; Hasegawa, S.; Nagai, H.; Uchida, F.; Yamatoji, M.; Kanno, N.I.; Yamagata, K.; Sakai, S.; Yanagawa, T.; Bukawa, H. MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J. Oral Pathol. Med. 2015, 45, 248–255. [Google Scholar] [CrossRef] [Green Version]
- Lerner, C.; Wemmert, S.; Bochen, F.; Kulas, P.; Linxweiler, M.; Hasenfus, A.; Heinzelmann, J.; Leidinger, P.; Backes, C.; Meese, E.; et al. Characterization of miR-146a and miR-155 in blood, tissue and cell lines of head and neck squamous cell carcinoma patients and their impact on cell proliferation and migration. J. Cancer Res. Clinic. Oncol. 2015, 142, 757–766. [Google Scholar] [CrossRef]
- Yang, C.-C.; Hung, P.-S.; Wang, P.-W.; Liu, C.-J.; Chu, T.-H.; Cheng, H.-W.; Lin, S.-C. miR-181 as a putative biomarker for lymph-node metastasis of oral squamous cell carcinoma. J. Oral Pathol. Med. 2011, 40, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.-H.; Bae, S.D.; Hong, H.S.; Kim, R.H.; Kang, M.K.; Park, N.-H. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem. Biophys. Res. Commun. 2011, 404, 896–902. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhou, X.; Li, S.; Jin, Y.; Chen, Z.; Chen, D.; Cai, Y.; Liu, Z.; Zhao, T.; Wang, A. MicroRNA-181a suppresses salivary adenoid cystic carcinoma metastasis by targeting MAPK–Snai2 pathway. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 5258–5266. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, A.C.; Scapulatempo-Neto, C.; Maia, D.C.; Evangelista, A.F.; Morini, M.A.; Carvalho, A.L.; Vettore, A.L. Accuracy of microRNAs as markers for the detection of neck lymph node metastases in patients with head and neck squamous cell carcinoma. BMC Med. 2015, 13, 108. [Google Scholar] [CrossRef] [Green Version]
- Arriagada, W.G.; Olivero, P.; Rodríguez, B.; Lozano-Burgos, C.; de Oliveira, C.E.; Coletta, R.D. Clinicopathological significance of miR-26, miR-107, miR-125b, and miR-203 in head and neck carcinomas. Oral Dis. 2018, 24, 930–939. [Google Scholar] [CrossRef]
- Obayashi, M.; Yoshida, M.; Tsunematsu, T.; Ogawa, I.; Sasahira, T.; Kuniyasu, H.; Imoto, I.; Abiko, Y.; Xu, D.; Fukunaga, S.; et al. microRNA-203 suppresses invasion and epithelial-mesenchymal transition induction via targeting NUAK1 in head and neck cancer. Oncotarget 2016, 7, 8223–8239. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-S.; Choi, D.W.; Kim, C.S.; Yu, S.-K.; Kim, H.-J.; Go, D.-S.; Lee, S.; Moon, S.M.; Kim, S.-G.; Chun, H.S.; et al. MicroRNA-203 Induces Apoptosis by TargetingBmi-1in YD-38 Oral Cancer Cells. Anticancer Res. 2018, 38, 3477–3485. [Google Scholar] [CrossRef]
- Yang, C.-J.; Shen, W.G.; Liu, C.-J.; Chen, Y.-W.; Lu, H.-H.; Tsai, M.-M.; Lin, S.-C. miR-221 and miR-222 expression increased the growth and tumorigenesis of oral carcinoma cells. J. Oral Pathol. Med. 2011, 40, 560–566. [Google Scholar] [CrossRef]
- Jiang, F.; Zhao, W.; Zhou, L.; Zhang, L.; Liu, Z.; Yu, D. miR-222 regulates the cell biological behavior of oral squamous cell carcinoma by targeting PUMA. Oncol. Rep. 2014, 31, 1255–1262. [Google Scholar] [CrossRef]
- Lopes, C.B.; Magalhães, L.; Teófilo, C.R.; Alves, A.P.N.N.; Montenegro, R.C.; Negrini, M.; Ribeiro-Dos-Santos, A. Differential expression of hsa-miR-221, hsa-miR-21, hsa-miR-135b, and hsa-miR-29c suggests a field effect in oral cancer. BMC Cancer 2018, 18, 721. [Google Scholar] [CrossRef]
- Liu, X.; Yu, J.; Jiang, L.; Wang, A.; Shi, F.; Ye, H.; Zhou, X. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genom. Proteom. 2009, 6, 131–139. [Google Scholar]
- Tachibana, H.; Sho, R.; Takeda, Y.; Zhang, X.; Yoshida, Y.; Narimatsu, H.; Otani, K.; Ishikawa, S.; Fukao, A.; Asao, H.; et al. Circulating miR-223 in Oral Cancer: Its Potential as a Novel Diagnostic Biomarker and Therapeutic Target. PLoS ONE 2016, 11, 159693. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lv, L.; Liu, X.; Jiang, X.; Yin, Q.; Hao, Y.; Xiao, L. MiR-223 promotes oral squamous cell carcinoma proliferation and migration by regulating FBXW7. Cancer Biomark. 2019, 24, 325–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, C.-H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef]
- Nohata, N.; Sone, Y.; Hanazawa, T.; Fuse, M.; Kikkawa, N.; Yoshino, H.; Chiyomaru, T.; Kawakami, K.; Enokida, H.; Nakagawa, M.; et al. miR-1 as a tumor suppressive microRNA targeting TAGLN2 in head and neck squamous cell carcinoma. Oncotarget 2011, 2, 29–42. [Google Scholar] [CrossRef] [Green Version]
- Nohata, N.; Hanazawa, T.; Kikkawa, N.; Sakurai, D.; Sasaki, K.; Chiyomaru, T.; Kawakami, K.; Yoshino, H.; Enokida, H.; Nakagawa, M.; et al. Identification of novel molecular targets regulated by tumor suppressive miR-1/miR-133a in maxillary sinus squamous cell carcinoma. Int. J. Oncol. 2011, 39, 1099–1107. [Google Scholar]
- Lu, J.; Zhao, F.P.; Peng, Z.; Zhang, M.W.; Lin, S.X.; Liang, B.; Zhang, B.; Liu, X.; Wang, L.; Li, G.; et al. EZH2 promotes angiogenesis through inhibition of miR-1/Endothelin-1 axis in nasopharyngeal carcinoma. Oncotarget 2014, 5, 11319–11332. [Google Scholar] [CrossRef]
- Peng, C.-Y.; Liao, Y.-W.; Lu, M.-Y.; Yu, C.-H.; Yu, C.-C.; Chou, M.-Y. Downregulation of miR-1 enhances tumorigenicity and invasiveness in oral squamous cell carcinomas. J. Formos. Med Assoc. 2017, 116, 782–789. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, J.; Chen, Z.; Wang, K.; Shi, L. MicroRNA-1-3p inhibits the proliferation and migration of oral squamous cell carcinoma cells by targeting DKK1. Biochem. Cell Boil. 2018, 96, 355–364. [Google Scholar] [CrossRef]
- Koshizuka, K.; Hanazawa, T.; Fukumoto, I.; Kikkawa, N.; Matsushita, R.; Mataki, H.; Mizuno, K.; Okamoto, Y.; Seki, N. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J. Hum. Genet. 2016, 62, 113–121. [Google Scholar] [CrossRef]
- Fukumoto, I.; Hanazawa, T.; Kinoshita, T.; Kikkawa, N.; Koshizuka, K.; Goto, Y.; Nishikawa, R.; Chiyomaru, T.; Enokida, H.; Nakagawa, M.; et al. MicroRNA expression signature of oral squamous cell carcinoma: Functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br. J. Cancer 2015, 112, 891–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, L.-F.; Wei, S.-B.; Gan, Y.-H.; Guo, Y.; Gong, K.; Mitchelson, K.; Cheng, J.; Yu, G.-Y. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int. J. Cancer 2014, 135, 2282–2293. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Chang, K.; Fan, C.; Zhang, Y. MiR-26a/miR-26b represses tongue squamous cell carcinoma progression by targeting PAK1. Cancer Cell Int. 2020, 20, 82–114. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, M.; Rao, A.K.D.M.; Arunkumar, G.; Rajkumar, K.S.; Rajaraman, R.; Munirajan, A.K. Down Regulation of miR-34a and miR-143 May Indirectly Inhibit p53 in Oral Squamous Cell Carcinoma: A Pilot Study. Asian Pac. J. Cancer Prev. 2015, 16, 7619–7625. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wang, X. MiR-34a targets BCL-2 to suppress the migration and invasion of sinonasal squamous cell carcinoma. Oncol. Lett. 2018, 16, 6566–6572. [Google Scholar] [CrossRef]
- Chen, Z.; Jin, Y.; Yu, N.; Wang, A.; Mahjabeen, I.; Wang, C.; Liu, X.; Zhou, X. Down-regulation of the microRNA-99 family members in head and neck squamous cell carcinoma. Oral Oncol. 2012, 48, 686–691. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Tymen, S.D.; Chen, D.; Fang, Z.J.; Zhao, Y.; Dragas, D.; Dai, Y.; Marucha, P.T.; Zhou, X. MicroRNA-99 Family Targets AKT/mTOR Signaling Pathway in Dermal Wound Healing. PLoS ONE 2013, 8, 64434. [Google Scholar] [CrossRef]
- Wang, Q.; Yan, B.; Fu, Q.; Lai, L.; Tao, X.; Fei, Y.; Shen, J.; Chen, Z. Downregulation of microRNA 99a in oral squamous cell carcinomas contributes to the growth and survival of oral cancer cells. Mol. Med. Rep. 2012, 6, 675–681. [Google Scholar] [CrossRef]
- Kuo, Y.-Z.; Tai, Y.-H.; Lo, H.-I.; Chen, Y.-L.; Cheng, H.-C.; Fang, W.-Y.; Lin, S.-H.; Yang, C.-L.; Tsai, S.-T.; Wu, L.-W. MiR-99a exerts anti-metastasis through inhibiting myotubularin-related protein 3 expression in oral cancer. Oral Dis. 2013, 20, 65–75. [Google Scholar] [CrossRef]
- He, K.; Tong, D.; Zhang, S.; Cai, D.; Wang, L.; Yang, Y.; Gao, L.; Chang, S.; Guo, B.; Song, T.; et al. miRNA-99b-3p functions as a potential tumor suppressor by targeting glycogen synthase kinase-3β in oral squamous cell carcinoma Tca-8113 cells. Int. J. Oncol. 2015, 47, 1528–1536. [Google Scholar] [CrossRef]
- Banerjee, R.; Mani, R.-S.; Russo, N.; Scanlon, C.S.; Tsodikov, A.; Jing, X.; Cao, Q.; Palanisamy, N.; Metwally, T.; Inglehart, R.C.; et al. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene 2011, 30, 4339–4349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, M.; Jiang, Y.-P.; Chen, W.; Li, K.-D.; Liu, X.; Gao, S.-Y.; Feng, H.; Wang, S.-S.; Jiang, J.; Ma, X.-R.; et al. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget 2015, 6, 6794–6810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.-R.; Wen, X.; He, Q.-M.; Li, Y.-Q.; Ren, X.-Y.; Yang, X.-J.; Zhang, J.; Wang, Y.-Q.; Ma, J.; Liu, N. MicroRNA-101 inhibits invasion and angiogenesis through targeting ITGA3 and its systemic delivery inhibits lung metastasis in nasopharyngeal carcinoma. Cell Death Dis. 2017, 8, 2566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kai, Y.; Peng, W.; Ling, W.; Jiebing, H.; Zhuan, B. Reciprocal effects between microRNA-140-5p and ADAM10 suppress migration and invasion of human tongue cancer cells. Biochem. Biophys. Res. Commun. 2014, 448, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Jing, P.; Sa, N.; Liu, X.; Liu, X.; Xu, W. MicroR-140-5p suppresses tumor cell migration and invasion by targeting ADAM10-mediated Notch1 signaling pathway in hypopharyngeal squamous cell carcinoma. Exp. Mol. Pathol. 2016, 100, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Pang, C. MicroRNA-140-5p inhibits the tumorigenesis of oral squamous cell carcinoma by targeting p21-activated kinase 4. Cell Boil. Int. 2019, 44, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Z.; Zou, Y.; Zhao, H. MicroRNA-140-5p inhibits salivary adenoid cystic carcinoma progression and metastasis via targeting survivin. Cancer Cell Int. 2019, 19, 301–312. [Google Scholar] [CrossRef]
- Yu, C.-C.; Chen, P.-N.; Peng, C.-Y.; Yu, C.-H.; Chou, M.-Y. Suppression of miR-204 enables oral squamous cell carcinomas to promote cancer stemness, EMT traits, and lymph node metastasis. Oncotarget 2016, 7, 20180–20192. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, F.; Zhou, X. miR-204-5p regulates cell proliferation and metastasis through inhibiting CXCR4 expression in OSCC. Biomed. Pharmacother. 2016, 82, 202–207. [Google Scholar] [CrossRef]
- Ma, L.; Deng, X.; Wu, M.; Zhang, G.; Huang, J. Down-regulation of miRNA-204 by LMP-1 enhances CDC42 activity and facilitates invasion of EBV-associated nasopharyngeal carcinoma cells. FEBS Lett. 2014, 588, 1562–1570. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhang, Y.; Zhou, D.; Cao, G.; Wu, Y. miR-204 enhances p27 mRNA stability by targeting Brd4 in head and neck squamous cell carcinoma. Oncol. Lett. 2018, 16, 4179–4184. [Google Scholar] [CrossRef] [PubMed]
- Libório-Kimura, T.N.; Jung, H.M.; Chan, E.K. miR-494 represses HOXA10 expression and inhibits cell proliferation in oral cancer. Oral Oncol. 2015, 51, 151–157. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Liao, X.; Yang, Q.; Liu, Y.; Peng, Y.; Zhong, H.; Yang, J.; Zhang, H.; Yu, Z.; Zuo, Y.; et al. MicroRNA-494-3p Promotes Cell Growth, Migration, and Invasion of Nasopharyngeal Carcinoma by Targeting Sox7. Technol. Cancer Res. Treat. 2018, 17, 1533033818809993. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, Y.; Lu, J.; Sun, Y.; Xiao, H.; Liu, M.; Tian, L. Combined detection of serum exosomal miR-21 and Hotair as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med. Oncol. 2014, 31, 148. [Google Scholar] [CrossRef] [PubMed]
- Hu, A.; Huang, J.-J.; Xu, W.-H.; Jin, X.-J.; Li, J.-P.; Tang, Y.-J.; Huang, X.-F.; Cui, H.-J.; Sun, G.-B. miR-21 and miR-375 microRNAs as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: Association with patient survival. Am. J. Transl. Res. 2014, 6, 604–613. [Google Scholar] [PubMed]
- Saito, K.; Inagaki, K.; Kamimoto, T.; Ito, Y.; Sugita, T.; Nakajo, S.; Hirasawa, A.; Iwamaru, A.; Ishikura, T.; Hanaoka, H.; et al. MicroRNA-196a Is a Putative Diagnostic Biomarker and Therapeutic Target for Laryngeal Cancer. PLoS ONE 2013, 8, e71480. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Chen, M.; Tao, Z.; Hua, Q.; Chen, S.; Xiao, B. Identification of predictive biomarkers for early diagnosis of larynx carcinoma based on microRNA expression data. Cancer Genet. 2013, 206, 340–346. [Google Scholar] [CrossRef]
- Wu, Y.; Yu, J.; Ma, Y.; Wang, F.; Liu, H. miR-148a and miR-375 may serve as predictive biomarkers for early diagnosis of laryngeal carcinoma. Oncol. Lett. 2016, 12, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Ayaz, L.; Gorur, A.; Yaroğlu, H.Y.; Ozcan, C.; Tamer, L. Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: potential early-detection markers for laryngeal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2013, 139, 1499–1506. [Google Scholar] [CrossRef]
- Xu, Y.; Lin, Y.P.; Yang, D.; Zhang, G.; Zhou, H.F. Expression of serum microRNA-378 and its clinical significance in laryngeal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 5137–5142. [Google Scholar]
- Grzelczyk, W.L.; Szemraj, J.; Kwiatkowska, S.; Jozefowicz-Korczynska, M. Serum expression of selected miRNAs in patients with laryngeal squamous cell carcinoma (LSCC). Diagn. Pathol. 2019, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Childs, G.; Fazzari, M.; Kung, G.; Kawachi, N.; Brandwein-Gensler, M.; McLemore, M.; Chen, Q.; Burk, R.D.; Smith, R.V.; Prystowsky, M.B.; et al. Low-Level Expression of MicroRNAs let-7d and miR-205 Are Prognostic Markers of Head and Neck Squamous Cell Carcinoma. Am. J. Pathol. 2009, 174, 736–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, H.; Hasegawa, S.; Uchida, F.; Terabe, T.; Kanno, N.I.; Kato, K.; Yamagata, K.; Sakai, S.; Kawashiri, S.; Sato, H.; et al. MicroRNA-205-5p suppresses the invasiveness of oral squamous cell carcinoma by inhibiting TIMP-2 expression. Int. J. Oncol. 2018, 52, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Avissar, M.; Christensen, B.C.; Kelsey, K.T.; Marsit, C.J. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin. Cancer Res. 2009, 15, 2850–2855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.-C.; Chang, J.T.-C.; Huang, Y.-C.; Huang, C.-C.; Chen, W.-H.; Lee, L.-Y.; Huang, B.-S.; Chen, Y.-J.; Li, H.-F.; Cheng, A.-J. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin. Biochem. 2015, 48, 115–121. [Google Scholar] [CrossRef]
- Philipone, E.; Yoon, A.J.; Wang, S.; Shen, J.; Ko, Y.C.K.; Sink, J.M.; Rockafellow, A.; Shammay, N.; Santella, R.M. MicroRNAs-208b-3p, 204-5p, 129-2-3p and 3065-5p as predictive markers of oral leukoplakia that progress to cancer. Am. J. Cancer Res. 2016, 6, 1537–1546. [Google Scholar] [PubMed]
- Zhang, X.-W.; Liu, N.; Chen, S.; Wang, Y.; Zhang, Z.-X.; Sun, Y.-Y.; Qiu, G.-B.; Fu, W.-N. High microRNA-23a expression in laryngeal squamous cell carcinoma is associated with poor patient prognosis. Diagn. Pathol. 2015, 10, 22. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Tian, L.; Ren, H.; Chen, X.; Wang, Y.; Ge, J.; Wu, S.; Sun, Y.; Liu, M.; Xiao, H. MicroRNA-101 is a potential prognostic indicator of laryngeal squamous cell carcinoma and modulates CDK8. J. Transl. Med. 2015, 13, 271. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Jia, S.; Xu, P. MicroRNA-9 as a novel prognostic biomarker in human laryngeal squamous cell carcinoma. Int. J. Clin. Exp. Med. 2014, 7, 5523–5528. [Google Scholar]
- Maia, D.C.; de Carvalho, A.C.; Horst, M.A.; Carvalho, A.L.; Scapulatempo-Neto, C.; Vettore, A.L. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J. Transl. Med. 2015, 13, 262. [Google Scholar] [CrossRef] [Green Version]
- Cappellesso, R.; Marioni, G.; Crescenzi, M.; Giacomelli, L.; Guzzardo, V.; Mussato, A.; Staffieri, A.; Martini, A.; Blandamura, S.; Fassina, A. The prognostic role of the epithelial-mesenchymal transition markers E-cadherin and Slug in laryngeal squamous cell carcinoma. Histopathology 2015, 67, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, A.M.; Heaford, A.C.; Trask, D.K. Detection of Metastatic Head and Neck Squamous Cell Carcinoma Using the Relative Expression of Tissue-Specific Mir-205. Transl. Oncol. 2008, 1, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, S.-C.; Liao, C.-T.; Peng, C.-H.; Cheng, A.-J.; Chen, S.-J.; Huang, C.-G.; Hsieh, W.-P.; Yen, T.-C. MicroRNAs MiR-218, MiR-125b, and Let-7g Predict Prognosis in Patients with Oral Cavity Squamous Cell Carcinoma. PLoS ONE 2014, 9, 102403. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Ishinaga, H.; Midorikawa, K.; Shah, S.A.; Nakamura, S.; Hiraku, Y.; Oikawa, S.; Murata, M.; Takeuchi, K. Circulating microRNAs as novel prognosis biomarkers for head and neck squamous cell carcinoma. Cancer Boil. Ther. 2015, 16, 1042–1046. [Google Scholar] [CrossRef]
- Sun, J.; Yong, J.; Zhang, H. MicroRNA-93, upregulated in serum of nasopharyngeal carcinoma patients, promotes tumor cell proliferation by targeting PDCD4. Exp. Ther. Med. 2020, 19, 2579–2587. [Google Scholar] [CrossRef] [Green Version]
- Bufalino, A.; Cervigne, N.K.; de Oliveira, C.E.; Fonseca, F.P.; Rodrigues, P.C.; Macedo, C.C.; Sobral, L.M.; Miguel, M.C.; Lopes, M.A.; Paes Leme, A.F.; et al. Low miR-143/miR-145 Cluster Levels Induce Activin a Overexpression in Oral Squamous Cell Carcinomas, Which Contributes to Poor Prognosis. PLoS ONE 2015, 10, 136599. [Google Scholar] [CrossRef]
- Sousa, L.O.; Sobral, L.; Matsumoto, C.S.; Saggioro, F.P.; López, R.V.; Panepucci, R.A.; Curti, C.; Silva, W.A.; Greene, L.J.; Leopoldino, A.M. Lymph node or perineural invasion is associated with low miR-15a, miR-34c and miR-199b levels in head and neck squamous cell carcinoma. BBA Clin. 2016, 6, 159–164. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Liu, L.; Fu, H.; Wang, Q.; Shi, Y. Association of Decreased Expression of Serum miR-9 with Poor Prognosis of Oral Squamous Cell Carcinoma Patients. Med. Sci. Monit. 2016, 22, 289–294. [Google Scholar] [CrossRef]
- Suh, Y.-E.; Raulf, N.; Gäken, J.; Lawler, K.; Urbano, T.G.; Bullenkamp, J.; Gobeil, S.; Huot, J.; Odell, E.; Tavassoli, M. MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. Int. J. Cancer 2015, 137, 1021–1034. [Google Scholar] [CrossRef]
- Liu, N.; Boohaker, R.J.; Jiang, C.; Boohaker, J.R.; Xu, B. A radiosensitivity MiRNA signature validated by the TCGA database for head and neck squamous cell carcinomas. Oncotarget 2015, 6, 34649–34657. [Google Scholar] [CrossRef]
- de Jong, M.C.; Hoeve, J.T.; Grenman, R.; Wessels, L.; Kerkhoven, R.; Riele, H.T.; Brekel, M.W.V.D.; Verheij, M.; Begg, A.C. Pretreatment microRNA expression impacting on epithelial to mesenchymal transition predicts intrinsic radiosensitivity in head and neck cancer celllines and patients. Clin. Cancer Res. 2015, 21, 5630–5638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Shi, L.; Lei, Y.-M.; Bao, Y.-H.; Li, Z.-Y.; Ding, F.; Zhu, G.-T.; Wang, Q.-Q.; Huang, C. Radiosensitization effect of hsa-miR-138-2-3p on human laryngeal cancer stem cells. PeerJ 2017, 5, 3233. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.-Y.; Hsieh, P.-L.; Wang, T.H.; Yu, C.-C.; Lu, M.-Y.; Liao, Y.-W.; Lee, T.-H.; Peng, C.-Y. Andrographolide impedes cancer stemness and enhances radio-sensitivity in oral carcinomas via miR-218 activation. Oncotarget 2016, 8, 4196–4207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, P.; Sana, J.; Slavik, M.; Gurin, D.; Radova, L.; Gablo, N.A.; Kazda, T.; Smilek, P.; Horakova, Z.; Gal, B.; et al. MicroRNA-15b-5p Predicts Locoregional Relapse in Head and Neck Carcinoma Patients Treated with Intensity-modulated Radiotherapy. Cancer Genom. Proteom. 2019, 16, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Yin, W.; Wang, P.; Wang, X.; Song, W.; Cui, X.; Yu, H.; Zhu, W. Identification of microRNAs and mRNAs associated with multidrug resistance of human laryngeal cancer Hep-2 cells. Braz. J. Med. Boil. Res. 2013, 46, 546–554. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Wang, J.; Huang, H.; Hou, J.; Zhang, B.; Wang, A. MiR-181a–Twist1 pathway in the chemoresistance of tongue squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2013, 441, 364–370. [Google Scholar] [CrossRef]
- Jiang, F.; Zhao, W.; Zhou, L.; Liu, Z.; Li, W.; Yu, D. MiR-222 Targeted PUMA to Improve Sensitization of UM1 Cells to Cisplatin. Int. J. Mol. Sci. 2014, 15, 22128–22141. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-Y.; Liu, Z.-J.; He, H.; Zhang, C.; Wang, Y.-L. MicroRNA-101-3p suppresses cell proliferation, invasion and enhances chemotherapeutic sensitivity in salivary gland adenoid cystic carcinoma by targeting Pim-1. Am. J. Cancer Res. 2015, 5, 3015–3029. [Google Scholar]
- Qin, X.; Guo, H.; Wang, X.; Zhu, X.; Yan, M.; Wang, X.; Xu, Q.; Shi, J.; Lu, E.; Chen, W.; et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Boil. 2019, 20, 12. [Google Scholar] [CrossRef]
- Kariya, A.; Furusawa, Y.; Yunoki, T.; Kondo, T.; Tabuchi, Y. A microRNA-27a mimic sensitizes human oral squamous cell carcinoma HSC-4 cells to hyperthermia through downregulation of Hsp110 and Hsp90. Int. J. Mol. Med. 2014, 34, 334–340. [Google Scholar] [CrossRef] [Green Version]
- Ruan, Q.; Fang, Z.; Cui, S.; Zhang, X.-L.; Wu, Y.-B.; Tang, H.-S.; Tu, Y.-N.; Ding, Y. Thermo-chemotherapy Induced miR-218 upregulation inhibits the invasion of gastric cancer via targeting Gli2 and E-cadherin. Tumor Boil. 2015, 36, 5807–5814. [Google Scholar] [CrossRef]
- Wald, A.I.; Hoskins, E.E.; Wells, S.I.; Ferris, R.; Khan, S.A. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck 2011, 33, 504–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lajer, C.B.; Garnæs, E.; Friis-Hansen, L.; Norrild, B.; Therkildsen, M.H.; Glud, M.; Rossing, M.; Lajer, H.; Svane, D.; Skotte, L.; et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: Bridging between HPV-related head and neck cancer and cervical cancer. Br. J. Cancer 2012, 106, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Mirghani, H.; Ugolin, N.; Ory, C.; Goislard, M.; Lefèvre, M.; Baulande, S.; Hofman, P.; Guily, J.L.S.; Chevillard, S.; Lacave, R. Comparative analysis of micro-RNAs in human papillomavirus-positive versus -negative oropharyngeal cancers. Head Neck 2016, 38, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Gay, H.A.; Chernock, R.D.; Zhang, T.R.; Luo, J.; Thorstad, W.L.; Lewis, J.S.; Wang, X. A microRNA expression signature for the prognosis of oropharyngeal squamous cell carcinoma. Cancer 2012, 119, 72–80. [Google Scholar] [CrossRef]
- Heß, J.; Unger, K.; Maihöfer, C.; Schüttrumpf, L.; Wintergerst, L.; Heider, T.; Weber, P.; Marschner, S.; Braselmann, H.; Samaga, D.; et al. A Five-MicroRNA Signature Predicts Survival and Disease Control of Patients with Head and Neck Cancer Negative for HPV Infection. Clin. Cancer Res. 2018, 25, 1505–1516. [Google Scholar] [CrossRef] [Green Version]
- Božinović, K.; Sabol, I.; Dediol, E.; Gašperov, N.M.; Manojlović, S.; Vojtechova, Z.; Tachezy, R.; Grce, M. Genome-wide miRNA profiling reinforces the importance of miR-9 in human papillomavirus associated oral and oropharyngeal head and neck cancer. Sci. Rep. 2019, 9, 2306. [Google Scholar] [CrossRef]
- Bersani, C.; Mints, M.; Tertipis, N.; Haeggblom, L.; Näsman, A.; Romanitan, M.; Dalianis, T.; Ramqvist, T. MicroRNA-155, -185 and -193b as biomarkers in human papillomavirus positive and negative tonsillar and base of tongue squamous cell carcinoma. Oral Oncol. 2018, 82, 8–16. [Google Scholar] [CrossRef]
- Amaral, N.S.D.; Melo, N.C.; Maia, B.D.M.; Rocha, R.M. Noncoding RNA Profiles in Tobacco and Alcohol-Associated Diseases. Genes 2016, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Momi, N.; Kaur, S.; Rachagani, S.; Ganti, A.K.; Batra, S.K.; Rachgani, S. Smoking and microRNA dysregulation: A cancerous combination. Trends Mol. Med. 2013, 20, 36–47. [Google Scholar] [CrossRef] [Green Version]
- Manikandan, M.; Rao, A.K.D.M.; Rajkumar, K.S.; Rajaraman, R.; Munirajan, A.K. Altered levels of miR-21, miR-125b-2, miR-138, miR-155, miR-184, and miR-205 in oral squamous cell carcinoma and association with clinicopathological characteristics. J. Oral Pathol. Med. 2014, 44, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Melling, G.; Hinsley, E.E.; Kabir, T.; Colley, H.E.; Murdoch, C.; Lambert, D.W. Cigarette smoke condensate promotes pro-tumourigenic stromal-epithelial interactions by suppressing miR-145. J. Oral Pathol. Med. 2012, 42, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Izzotti, A.; Calin, G.A.; Arrigo, P.; Steele, V.E.; Croce, C.M.; de Flora, S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2008, 23, 806–812. [Google Scholar] [CrossRef] [PubMed]
- House, R.; Majumder, M.; Janakiraman, H.; Ogretmen, B.; Kato, M.; Erkul, E.; Hill, E.; Atkinson, C.; Barth, J.; Day, T.A.; et al. Smoking-induced control of miR-133a-3p alters the expression of EGFR and HuR in HPV-infected oropharyngeal cancer. PLoS ONE 2018, 13, e0205077. [Google Scholar] [CrossRef] [Green Version]
- Gong, S.-Q.; Xu, M.; Xiang, M.-L.; Shan, Y.-M.; Zhang, H. The Expression and Effection of MicroRNA-499a in High-Tobacco Exposed Head and Neck Squamous Cell Carcinoma: A Bioinformatic Analysis. Front. Oncol. 2019, 9, 678. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Yokota, S.-I.; Tatsumi, N.; Fukami, T.; Yokoi, T.; Nakajima, M. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol. Appl. Pharmacol. 2013, 272, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Yamada, H.; Nagura, A.; Ohashi, K.; Ishikawa, K.; Yamazaki, M.; Ando, Y.; Ichino, N.; Osakabe, K.; Sugimoto, K.; et al. Association of cigarette smoking with serum microRNA expression among middle-aged Japanese adults. Fujita Med. J. 2016, 2. [Google Scholar]
- Saad, M.A.; Kuo, S.Z.; Rahimy, E.; Zou, A.E.; Korrapati, A.; Rahimy, M.; Kim, E.; Zheng, H.; Yu, M.A.; Wang-Rodriguez, J.; et al. Alcohol-dysregulated miR-30a and miR-934 in head and neck squamous cell carcinoma. Mol. Cancer 2015, 14, 181. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Liu, H. MicroRNA-10a-5p and microRNA-34c-5p in laryngeal epithelial premalignant lesions: Differential expression and clinicopathological correlation. Eur. Arch. Oto-Rhino-Laryngol. 2014, 272, 391–399. [Google Scholar] [CrossRef]
- Supic, G.; Zeljic, K.; Rankov, A.D.; Kozomara, R.; Nikolic, A.; Radojkovic, D.; Magic, Z. miR-183 and miR-21 expression as biomarkers of progression and survival in tongue carcinoma patients. Clin. Oral Investig. 2017, 22. [Google Scholar] [CrossRef]
- Krishnan, A.R.; Zheng, H.; Kwok, J.G.; Qu, Y.; Zou, A.E.; Korrapati, A.; Li, P.X.; Califano, J.; Hovell, M.F.; Wang-Rodriguez, J.; et al. A comprehensive study of smoking-specific microRNA alterations in head and neck squamous cell carcinoma. Oral Oncol. 2017, 72, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, M.; Rao, A.K.D.M.; Arunkumar, G.; Manickavasagam, M.; Rajkumar, K.S.; Rajaraman, R.; Munirajan, A.K. Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol. Cancer 2016, 15, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricciardiello, F.; Capasso, R.; Kawasaki, H.; Abate, T.; Oliva, F.; Lombardi, A.; Misso, G.; Ingrosso, D.; Leone, C.; Iengo, M.; et al. A miRNA signature suggestive of nodal metastases from laryngeal carcinoma. Acta Otorhinolaryngol. Ital. 2017, 37, 467–474. [Google Scholar] [PubMed]

| Cancer Type | miRNA | Regulation | Role | Target Gene | Ref. |
|---|---|---|---|---|---|
| Laryngeal cancer (LCa) | Let-7a | Down | Tumor-suppressive | HMGA2 | [43] |
| miR-1 | Down | Tumor-suppressive | FN1 | [25,35] | |
| miR-9 | Up | Oncogenic | PTEN | [23] | |
| miR-9 | Up | Oncogenic | - | [109] | |
| miR-10b | Up | Oncogenic | CDH1 | [24] | |
| miR-21 | Up | Oncogenic | BTG2, PTEN, TPM1, PDCD4, CDK2AP1 | [25] | |
| miR-23a | Up | Oncogenic | - | [107] | |
| miR-24 | Down | Tumor-suppressive | XIAP | [36] | |
| miR-34a | Down | Tumor-suppressive | CCND1, GALNT7 | [38,40] | |
| miR-34c | Down | Tumor-suppressive | c-Met, GALNT7 | [39,40] | |
| miR-93 | Up | Oncogenic | CCNG2 | [28,29] | |
| miR-101 | Down | Tumor-suppressive | CDK8 | [108] | |
| miR-126 | - | Tumor-suppressive | Camsap1 | [30] | |
| miR-132 | Up | Oncogenic | FOXO1 | [31] | |
| miR-138-2-3p | Up | Oncogenic | - | [122] | |
| miR-144 | Down | Tumor-suppressive | IRS1 | [41] | |
| miR-203 | Down | Tumor-suppressive | - | [121] | |
| miR-205 | Up | Oncogenic | CDK2AP1 | [27] | |
| miR-221 | - | Oncogenic | ER-α, p27, p57, c-kit, Apaf-1 | [32] | |
| miR-296-5p | Up | Oncogenic | - | [110] | |
| miR-302-3p | Up | Oncogenic | Smad4 | [33] | |
| miR-340 | Down | Tumor-suppressive | p27, EZH2 | [42] | |
| miR-423-3p | Up | Oncogenic | AdipoR2 | [34] | |
| Head and neck squamous cell carcinoma (HNSCC) | miR-1 | Down | Tumor-suppressive | TAGLN2 | [65] |
| miR-1 | Down | Tumor-suppressive | EGFR, c-Met | [70] | |
| miR-10b | Down | Tumor-suppressive | - | [45] | |
| miR-99 | Down | Tumor-suppressive | IGF1R, mTOR, Akt1 | [76,77] | |
| miR-100 | Down | Tumor-suppressive | IGF1R, mTOR, Akt1 | [76,77] | |
| miR-101 | Down | Tumor-suppressive | EZH2, rap1GAP | [81,82] | |
| miR-146a | Down | Tumor-suppressive | - | [50] | |
| miR-155 | Down | Tumor-suppressive | - | [50] | |
| miR-196a | Up | Oncogenic | - | [45] | |
| miR-203 | Up | Oncogenic | - | [54] | |
| miR-203 | Down | Tumor-suppressive | - | [56] | |
| miR-204 | Down | Tumor-suppressive | Brd4 | [91] | |
| miR-205 | Up | Oncogenic | - | [54] | |
| miR-206 | Down | Tumor-suppressive | EGFR, c-Met | [70] | |
| Oral squamous cell cancer (OSCC) | miR-1 | Down | Tumor-suppressive | Slug | [68] |
| miR-1-3p | Down | Tumor-suppressive | DKK1 | [69] | |
| miR-26a/b | Down | Tumor-suppressive | TMEM184B | [71] | |
| miR-34a | Down | Tumor-suppressive | - | [74] | |
| miR-99-3b | Down | Tumor-suppressive | glycogen synthase kinase-3β | [80] | |
| miR-99a | Down | Tumor-suppressive | Myotubularin-related protein 3 | [78,79] | |
| miR-104-5p | Down | Tumor-suppressive | PAK4 | [86] | |
| miR-155 | Up | Oncogenic | - | [141] | |
| miR-155 | Up | Oncogenic | CDC73 | [47,48,49] | |
| miR-181a | Down | Tumor-suppressive | K-ras | [52] | |
| miR-181a/b | Up | Oncogenic | - | [51] | |
| miR-204 | Down | Tumor-suppressive | Sox4, Slug | [88] | |
| miR-204-5p | Down | Tumor-suppressive | CXCR4 | [89] | |
| miR-222 | Up | Oncogenic | PUMA | [60] | |
| miR-222 | Down | Tumor-suppressive | PUMA | [127] | |
| miR-223 | Up | Oncogenic | - | [62] | |
| miR-223 | Up | Oncogenic | FBXW7 | [63] | |
| miR-494 | Down | Tumor-suppressive | HOXA10 | [90] | |
| Oral | miR-10b | Up | Oncogenic | - | [44] |
| Oral tongue squamous cell carcinoma (OTSCC) | miR-222 | Down | Tumor-suppressive | MMP1, SOD2 | [61] |
| Hypopharyngeal squamous cell carcinoma (HSCC) | miR-140-5p | Down | Tumor-suppressive | ADAM10 | [85] |
| Maxillary sinus | miR-1 | Down | Tumor-suppressive | TAGLN2, PNP | [66] |
| Nasopharyngeal carcinoma (NPC) | miR-1 | Down | Tumor-suppressive | ET-1 | [67] |
| miR-10b | Down | Tumor-suppressive | - | [46] | |
| miR-101 | Down | Tumor-suppressive | ITGA3 | [83] | |
| miR-204 | Down | Tumor-suppressive | Cdc42 | [90] | |
| miR-494-3p | Up | Oncogenic | Sox7 | [93] | |
| Salivary adenoid cystic carcinoma (SACC) | miR-104-5p | Down | Tumor-suppressive | Survivin | [87] |
| miR-181a | Down | Tumor-suppressive | MAP2K1, MAPK1, Snai2 | [53] | |
| Salivary gland adenoid cystic carcinoma (SGACC) | miR-101-3p | Down | Tumor-suppressive | Pim-1 | [128] |
| Sinonasal squamous cell carcinomas (SN-SCC) | miR-34a | Down | Tumor-suppressive | BCL-2 | [75] |
| Tongue | miR-21 | Up | Oncogenic | - | [148] |
| miR-183 | Up | Oncogenic | - | [148] | |
| Tongue squamous cell carcinoma (TSCC) | miR-26a | Down | Tumor-suppressive | DNMT3B | [72] |
| miR-26a | Down | Tumor-suppressive | PAK1 | [73] | |
| miR-26b | Down | Tumor-suppressive | PAK1 | [73] | |
| miR-140-5p | - | Tumor-suppressive | LAMC1, HDAC7, PAX6 | [84] | |
| miR-181a | Down | Tumor-suppressive | Twist1 | [126] |
| miRNA | Regulation | Cancer | Diagnosis/Prognosis | Ref. |
|---|---|---|---|---|
| Let-7a | Up | LCa | Diagnosis | [101] |
| Let-7g | Up | OSCC | Prognosis | [113] |
| miR-100 | Down | HNSCC | Prognosis | [117] |
| miR-101 | Down | LCa | Prognosis | [108] |
| miR-10b | Up | OSCC | Diagnosis | [44] |
| miR-125b | Up | OSCC | Prognosis | [113] |
| miR-125b | Down | HNSCC | Prognosis | [117] |
| miR-1287 | Down | LCa | Diagnosis | [97] |
| miR-1303 | Up | LCa | Diagnosis | [99] |
| miR-133b | Down | LCa | Diagnosis | [96] |
| miR-143 | Down | OSCC | Prognosis | [116] |
| miR-145 | Down | OSCC | Prognosis | [116] |
| miR-146a | Down | HNSCC | Diagnosis | [50] |
| miR-155 | Up | OSCC | Prognosis | [48,49] |
| miR-15a | Down | HNSCC | Diagnosis, Prognosis | [117] |
| miR-181a | Down | SACC, TSCC | Prognosis | [53,126] |
| miR-196a | Up | LCa, OSCC | Diagnosis | [96,105] |
| miR-196b | Up | OSCC | Diagnosis | [105] |
| miR-199b | Down | HNSCC | Diagnosis, Prognosis | [117] |
| miR-200a | Down | LCa | Prognosis | [111] |
| miR-200c | Down | LCa | Prognosis | [111] |
| miR-203 | Up | HNSCC | Prognosis | [54] |
| miR-203 | Down | HNSCC | Prognosis | [121] |
| miR-204 | Down | OSCC | Prognosis | [88] |
| miR-205 | Down | HNSCC | Prognosis | [102] |
| miR-205 | Up | HNSCC | Prognosis | [112] |
| miR-205-5p | Down | OSCC | Diagnosis, Prognosis | [103] |
| miR-21 | Up | HNSCC | Prognosis | [114,117] |
| miR-21 | Up | LCa | Diagnosis, Prognostic | [94,95] |
| miR-212-3p | Up | LCa | Diagnosis | [99] |
| miR-218 | Up | OSCC | Prognosis | [113] |
| miR-223 | Up | HNSCC | Prognosis | [114] |
| miR-23a | Up | LCa | Prognosis | [107] |
| miR-296-5p | Up | LCa | Prognosis | [110] |
| miR-31 | Up | LCa | Diagnosis | [101] |
| miR-33 | Up | LCa | Diagnosis | [101] |
| miR-331-3p | Up | LCa | Diagnosis | [99] |
| miR-34c | Down | HNSCC | Diagnosis, Prognosis | [117] |
| miR-375 | Down | LCa | Diagnosis, Prognosis | [95] |
| miR-375 | Down | LCa | Diagnosis | [98] |
| miR-378 | Up | LCa | Diagnosis | [100] |
| miR-455-5p | Up | LCa | Diagnosis | [96] |
| miR-603 | Up | LCa | Diagnosis | [99] |
| miR-657 | Up | LCa | Diagnosis | [97] |
| miR-9 | Up | LCa | Prognosis | [109] |
| miR-9 | Down | OSCC | Prognosis | [118] |
| miR-93 | Up | HNSCC | Prognosis | [28,115] |
| miR-99a | Down | HNSCC | Prognosis | [114] |
| miRNA | Cancer | Sample Type | Combinatorial Treatment | Effect | Ref. |
|---|---|---|---|---|---|
| miR-24 | LCa | LSCC cell lines (Hep-2, AMC-HN-8) | Overexpressed miR-24 (pGCMV/miR-24) + Radiation | Enhanced radiosensitivity: Suppresses cell proliferation and induces cell apoptosis | [36] |
| miR-196a | HNSCC | HNC cell line (HN30) | miR-196a knockdown (miR-196a sponge plasmid) + Radiation | Enhanced radiosensitivity: Decreases cell viability | [119] |
| miR-138-2-3p | LCa | LCa stem cells (originated from Hep-2) | Overexpressed miR-138-2-3p (100 nM miR-138-2-3p mimic) + Radiation | Enhanced radiosensitivity: Inhibits cell proliferation, viability, invasion and induces cell apoptosis, cell cycle arrest and DNA damage | [122] |
| miR-218 | Oral cancer | Oral cancer stem cells | Overexpressed miR-218 (pLV-miR-218) + Radiation | Enhanced radiosensitivity: Decreases cell viability | [123] |
| miR-181a | TSCC | DDP-resistant TSCC cell line (originated from CAL27) | Overexpressed miR-181a (miR-181a mimic) + Cisplatin | Enhanced chemosensitivity: Decreases IC50 value to cisplatin | [126] |
| miR-222 | OSCC | OSCC cell line (UM1) | miR-222 knockdown (anti-miR-222) + Cisplatin | Enhanced chemosensitivity: Induces cell apoptosis by up-regulation of pro-apoptotic PUMA expression and reduces cell invasiveness and IC50 value to cisplatin | [127] |
| miR-101-3p | SGACC | SGACC cell lines (SACC-LM, SACC-83) | miR-101-3p knockdown (anti-miR-101-3p) + Cisplatin | Enhanced chemosensitivity: Inhibits cell growth and induces cell apoptosis | [128] |
| miR-196a | HNC | HNC cell line (CAL27) | miR-196a knockdown (anti-miR-196a) + Cisplatin | Enhanced chemosensitivity: Promotes cell apoptosis and decreases colony formation | [129] |
| miRNA | Regulation | Cancer | Resistance | Target Gene | Ref. |
|---|---|---|---|---|---|
| let-7e | Down | HNSCC | Radiation | - | [120] |
| miR-101-3p | Down | SGACC | Chemotherapy | Pim-1 | [128] |
| miR-1254 | Up | HNSCC | Radiation | - | [120] |
| miR-138-2-3p | Up | LCa | Radiation | - | [122] |
| miR-150 | Up | HNSCC | Radiation | - | [120] |
| miR-16 | Up | HNSCC | Radiation | - | [120] |
| miR-181a | Down | TSCC | Chemotherapy | Twist1 | [126] |
| miR-183-star | Up | LCa | Radiation | - | [110] |
| miR-196a | Up | HNSCC | Radiation | Annexin A1 | [119] |
| miR-196b | Up | HNC | Chemotherapy | CDKN1B, ING5 | [129] |
| miR-200c | Up | LCa | Radiation | - | [110] |
| miR-203 | Down | LCa | Radiation | - | [121] |
| miR-203 | Down | OSCC | Thermotherapy | - | [130] |
| miR-210 | Up | LCa | Chemotherapy | - | [125] |
| miR-218 | - | OSCC | Radiation | Bim1 | [123] |
| miR-222 | Down | OSCC | Chemotherapy | PUMA | [127] |
| miR-23a | Up | OSCC | Thermotherapy | - | [130] |
| miR-24 | Down | LCa | Radiation | XIAP | [36] |
| miR-25-star | Down | LCa | Chemotherapy | - | [125] |
| miR-27a | Up | OSCC | Thermotherapy | - | [130] |
| miR-296-5p | Up | LCa | Radiation | - | [110] |
| miR-29b | Up | HNSCC | Radiation | - | [120] |
| miR-30a | Down | OSCC | Thermotherapy | - | [130] |
| miR-30c | Down | OSCC | Thermotherapy | - | [130] |
| miR-424 | Down | LCa | Chemotherapy | - | [125] |
| miR-452 | Up | LCa | Radiation | - | [110] |
| miR-494 | Down | LCa | Chemotherapy | - | [125] |
| miR-923 | Up | LCa | Chemotherapy | - | [125] |
| miR-93 | Down | LCa | Chemotherapy | - | [125] |
| miR-93-star | Down | LCa | Chemotherapy | - | [125] |
| miRNA | Regulation | Region | Infection or Habit | Ref. |
|---|---|---|---|---|
| miR-101 | Up | HNSCC | Alcohol | [148] |
| miR-107 | Down | Oropharyngeal SCC | HPV | [134] |
| miR-1266 | Up | HNSCC | Alcohol | [148] |
| miR-133a-3p | Down | Oropharyngeal SCC | Smoking | [144] |
| miR-139-5p | Down | HNSCC | HPV | [133] |
| miR-142-5p | Down | HNSCC | HPV | [132] |
| miR-143 | Down | HNSCC | HPV | [133] |
| miR-145 | Down | HNSCC | HPV | [133] |
| miR-145 | Down | Oral Fibroblast | Smoking | [142] |
| miR-155 | Down | HNSCC | HPV | [132] |
| miR-155 | Up | Oropharyngeal | HPV | [135] |
| miR-155 | Up | OSCC | Smoking | [141] |
| miR-155 | Up | TSCC/BOTSCC | HPV | [138] |
| miR-15a | Up | HNSCC | HPV | [133] |
| miR-16 | Up | HNSCC | HPV | [133] |
| miR-181a | Down | HNSCC | HPV | [132] |
| miR-181b | Down | HNSCC | HPV | [132] |
| miR-183 | Up | Tongue Cancer | Alcohol | [150] |
| miR-18a | Down | Oropharyngeal SCC | HPV | [135] |
| miR-195 | Down | HNSCC | HPV | [133] |
| miR-218 | Down | HNSCC | HPV | [132] |
| miR-221 | Down | HNSCC | HPV | [132] |
| miR-222 | Down | HNSCC | HPV | [132] |
| miR-223 | Down | Oropharyngeal SCC | HPV | [135] |
| miR-29a | Down | HNSCC | HPV | [132] |
| miR-30a | Up | HNSCC | Alcohol | [148] |
| miR-31 | Down | Oropharyngeal SCC | HPV | [135] |
| miR-3164 | Up | HNSCC | Alcohol | [148] |
| miR-3178 | Up | HNSCC | Alcohol | [148] |
| miR-324-5p | Down | Oropharyngeal SCC | HPV | [134] |
| miR-33 | Up | HNSCC | HPV | [132] |
| miR-34a | Up | OSCC | Alcohol | [74] |
| miR-34c-5p | Up | Laryngeal Epithelial Premalignant Lesions | Alcohol | [149] |
| miR-363 | Up | HNSCC | HPV | [132] |
| miR-3690 | Up | HNSCC | Alcohol | [148] |
| miR-381 | Down | HNSCC | HPV | [133] |
| miR-497 | Up | HNSCC | HPV | [132] |
| miR-497 | Down | HNSCC | HPV | [133] |
| miR-499a | Up | HNSCC | Smoking | [145] |
| miR-574-3p | Down | HNSCC | HPV | [133] |
| miR-675 | Up | HNSCC | Alcohol | [148] |
| miR-9 | Up | Oropharyngeal SCC | HPV | [135] |
| miR-9 | Up | OSCC, Oropharyngeal SCC | HPV | [137] |
| miR-934 | Up | HNSCC | Alcohol | [148] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takeuchi, T.; Kawasaki, H.; Luce, A.; Cossu, A.M.; Misso, G.; Scrima, M.; Bocchetti, M.; Ricciardiello, F.; Caraglia, M.; Zappavigna, S. Insight toward the MicroRNA Profiling of Laryngeal Cancers: Biological Role and Clinical Impact. Int. J. Mol. Sci. 2020, 21, 3693. https://doi.org/10.3390/ijms21103693
Takeuchi T, Kawasaki H, Luce A, Cossu AM, Misso G, Scrima M, Bocchetti M, Ricciardiello F, Caraglia M, Zappavigna S. Insight toward the MicroRNA Profiling of Laryngeal Cancers: Biological Role and Clinical Impact. International Journal of Molecular Sciences. 2020; 21(10):3693. https://doi.org/10.3390/ijms21103693
Chicago/Turabian StyleTakeuchi, Takashi, Hiromichi Kawasaki, Amalia Luce, Alessia Maria Cossu, Gabriella Misso, Marianna Scrima, Marco Bocchetti, Filippo Ricciardiello, Michele Caraglia, and Silvia Zappavigna. 2020. "Insight toward the MicroRNA Profiling of Laryngeal Cancers: Biological Role and Clinical Impact" International Journal of Molecular Sciences 21, no. 10: 3693. https://doi.org/10.3390/ijms21103693
APA StyleTakeuchi, T., Kawasaki, H., Luce, A., Cossu, A. M., Misso, G., Scrima, M., Bocchetti, M., Ricciardiello, F., Caraglia, M., & Zappavigna, S. (2020). Insight toward the MicroRNA Profiling of Laryngeal Cancers: Biological Role and Clinical Impact. International Journal of Molecular Sciences, 21(10), 3693. https://doi.org/10.3390/ijms21103693