Identifying and Remediating Soil Microbial Legacy Effects of Invasive Grasses for Restoring California Coastal Sage Scrub Ecosystems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Site
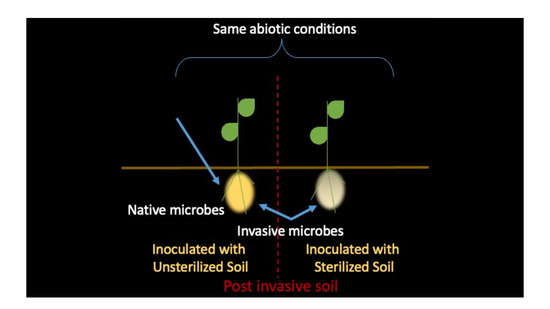
2.2. Experimental Setup
2.3. Soil Inoculum
2.4. Plant Growth Conditions
2.5. Plant Harvest
2.6. Soil Core Collection
2.7. Leachate Analyses
2.8. 16S DNA Extraction Quantification and Barcoded Targeted Amplicon Sequencing
2.9. AMF DNA Extraction Quantification and Barcoded Targeted Amplicon Sequencing
2.10. DNA Extraction from Roots
2.11. Data Analysis
3. Results
3.1. Bacteria and Leachate
3.2. Rhizosphere by Treatment
3.3. Fungal Composition
3.4. Plant Growth Trends
4. Discussion
4.1. Bacterial Composition
4.1.1. Roots and Rhizosphere
4.1.2. Rhizosphere by Treatment
4.2. Fungal Composition
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DiTomaso, J.M. Invasive weeds in rangelands: Species, impacts, and management. Weed Sci. 2000, 48, 255–265. [Google Scholar]
- Westman, E.W. Diversity relations and succession in Californian coastal sage scrub. Ecology 1981, 62, 170–184. [Google Scholar] [CrossRef]
- Rubinoff, D. Evaluating the California Gnatcatcher as an umbrella species for conservation of southern California coastal sage scrub. Conserv. Biol. 2001, 15, 1374–1383. [Google Scholar]
- D’Antonio, C.M.; Vitousek, P.M. Biological Invasions by Exotic Grasses, the Grass/Fire Cycle, and Global Change. Annu. Rev. Ecol. Syst. 1992, 23, 63–87. [Google Scholar] [CrossRef]
- Lowry, E.; Rollinson, E.J.; Laybourn, A.J.; Scott, T.E.; Aiello-Lammens, M.E.; Gray, S.M.; Mickley, J.; Gurevitch, J. Biological invasions: A field synopsis, systematic review, and database of the literature. Ecol. Evol. 2012, 3, 182–196. [Google Scholar] [CrossRef]
- Eviner, V.T.; Hoskinson, S.A.; Hawkes, C.V. Ecosystem impacts of exotic plants can feed back to increase invasion in western US rangelands. Rangelands 2010, 32, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Ehrenfeld, J.G. Effects of exotic plant invasions on soil nutrient cycling processes. Ecosystems 2003, 6, 503–523. [Google Scholar]
- Saggar, S.; McIntosh, P.D.; Hedley, C.B.; Knicker, H. Changes in soil microbial biomass, metabolic quotient, and organic matter turnover under Hieracium (H. pilosella L.). Biol. Fertil. Soils 1999, 30, 232–238. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G.; Ravit, B.; Elgersma, K. Feedback in the Plant-Soil System. Ann. Rev. Environ. Resour. 2005, 30, 75–115. [Google Scholar] [CrossRef] [Green Version]
- Dickens, S.J.M.; Allen, E.B.; Santiago, L.; Crowley, D. Exotic annuals reduce soil heterogeneity in coastal sage scrub soil chemical and biological characteristics. Soil Biol. Biochem. 2013, 58, 70–81. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, T.; Choi, B.; Kim, Y.; Kim, E. Invasive Lactuca serriola seeds contain endophytic bacteria that contribute to drought tolerance. Sci. Rep. 2021, 11, 13307. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Pei, Y.; Huang, W.; Ding, J.; Siemann, E. Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J. 2021, 15, 1919–1930. [Google Scholar] [PubMed]
- McLeod, M.L.; Bullington, L.; Cleveland, C.C.; Rousk, J.; Lekberg, Y. Invasive plant-derived dissolved organic matter alters microbial communities and carbon cycling in soils. Soil Biol. Biochem. 2021, 156, 108191. [Google Scholar] [CrossRef]
- Bever, J.D.; Schultz, P.A.; Pringle, A.; Morton, J.B. Arbuscular Mycorrhizal Fungi: More Diverse than Meets the Eye, and the Ecological Tale of Why. Bioscience 2001, 51, 923–931. [Google Scholar] [CrossRef] [Green Version]
- Batten, K.M.; Scow, K.M.; Davies, K.F.; Harrison, S.P. Two Invasive Plants Alter Soil Microbial Community Composition in Serpentine Grasslands. Biol. Invasions 2006, 8, 217–230. [Google Scholar] [CrossRef]
- Lankau, E.W.; Lankau, R.A. Plant species capacity to drive soil fungal communities contributes to differential impacts of plant–soil legacies. Ecology 2014, 95, 3221–3228. [Google Scholar] [CrossRef]
- Miller, R.M.; Jastrow, J.D. Mycorrhizal fungi influence soil structure. In Arbuscular Mycorrhizas: Molecular Biology and Physiology; Kluwer Academic Press: Dordrecht, The Netherlands, 2000; pp. 3–18. [Google Scholar]
- Klironomos, J. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 2002, 417, 67–70. [Google Scholar] [CrossRef]
- Barto, K.; Friese, C.; Cipollini, D. Arbuscular Mycorrhizal Fungi Protect a Native Plant from Allelopathic Effects of an Invader. J. Chem. Ecol. 2010, 36, 351–360. [Google Scholar] [CrossRef]
- Vogelsang, K.M.; Bever, J.D.; Griswold, M.; Schultz, P.A. The Use of Mycorrhizal Fungi in Erosion Control Applications; Final Report for Caltrans Contract No. 65A0070; California Department of Transportation: Sacramento, CA, USA, 2004. [Google Scholar]
- Aprahamian, A.M.; Lulow, M.E.; Major, M.R.; Balazs, K.R.; Treseder, K.K.; Maltz, M.R. Arbuscular mycorrhizal inoculation in coastal sage scrub restoration. Botany 2016, 94, 493–499. [Google Scholar] [CrossRef]
- Sieverding, E.; Oehl, F. Are arbuscular mycorrhizal fungal species invasive-derived from our knowledge about their distribution in different ecosystems? In BCPC Symposium Proceedings No. 81: Plant Protection and Plant Health in Europe: Introduction and Spread of Invasive Species; Monograph Series; The British Crop Protection Council: Alton, UK, 2005; pp. 197–202. [Google Scholar]
- Bever, J.D.; Dickie, I.A.; Facelli, E.; Facelli, J.M.; Klironomos, J.; Moora, M.; Rillig, M.C.; Stock, W.D.; Tibbett, M.; Zobel, M. Rooting theories of plant community ecology in microbial interactions. Trends Ecol. Evol. 2010, 25, 468–478. [Google Scholar]
- Cuddington, K. Legacy effects: The persistent impact of ecological interactions. Biol. Theory 2011, 6, 203–210. [Google Scholar] [CrossRef]
- Hawkes, C.V.; Belnap, J.; D’Antonio, C.; Firestone, M.K. Arbuscular mycorrhizal assemblages in native plant roots change in the presence of invasive exotic grasses. Plant Soil 2006, 281, 369–380. [Google Scholar] [CrossRef]
- Jordan, N.R.; Larson, D.L.; Huerd, S.C. Soil modification by invasive plants: Effects on native and invasive species of mixed-grass prairies. Biol. Invasions 2008, 10, 177–190. [Google Scholar]
- Mangla, S.; Callaway, I.M.; Callaway, R.M. Exotic invasive plant accumulates native soil pathogens which inhibit native plants. J. Ecol. 2008, 96, 58–67. [Google Scholar] [CrossRef]
- Liao, C.; Peng, R.; Luo, Y.; Zhou, X.; Wu, X.; Fang, C.; Chen, J.; Li, B. Altered ecosystem carbon and nitrogen cycles by plant invasion: A meta-analysis. New Phytol. 2008, 177, 706–714. [Google Scholar] [PubMed]
- Belnap, J.; Phillips, S.L.; Sherrod, S.K.; Moldenke, A. Soil biota can change after exotic plant invasion: Does this affect ecosystem processes? Ecology 2005, 86, 3007–3017. [Google Scholar]
- Bozzolo, F.H.; Lipson, D.A. Differential responses of native and exotic coastal sage scrub plant species to N additions and the soil microbial community. Plant Soil 2013, 371, 37–51. [Google Scholar] [CrossRef]
- Tran, J.; Cavagnaro, T.R. Growth and mycorrhizal colonization of two grasses in soils with different inundation histories. J. Arid Environ. 2010, 74, 715–717. [Google Scholar] [CrossRef]
- Asghari, H.; Cavagnaro, T. Arbuscular mycorrhizas enhance plant interception of leached nutrients. Funct. Plant Biol. 2011, 38, 219–226. [Google Scholar]
- Calflora: Information on California Plants for Education, Research, and Conservation. The Calflora Database. Distribution Data from Eileen Berbeo and Tarja Sag, Council for Watershed Health, and Cal-IPC Checklists. 2014. Available online: http://www.calflora.org/cgi-bin/noccdetail.cgi?seq_num=wb830-264andtaxon=Phalaris+aquatica (accessed on 11 May 2018).
- Bowler, P.A. Ecological restoration of coastal sage scrub and its potential role in habitat conservation plans. Environ. Manag. 2000, 26, S85–S96. [Google Scholar] [CrossRef]
- Weatherburn, M.W. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 1967, 39, 971–974. [Google Scholar]
- Doane, T.A.; Horwath, W.R. Spectrophotometric determination of nitrate with a single reagent. Anal. Lett. 2003, 36, 2713–2722. [Google Scholar]
- Pickett, B.; Irvine, I.C.; Bullock, E.; Arogyaswamy, K.; Aronson, E. Legacy effects of invasive grass impact soil microbes and native shrub growth. Invasive Plant Sci. Manag. 2019, 12, 22–35. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [PubMed]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI biosystems database. Nucleic Acids Res. 2010, 38, 492–496. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, K.T.; Maltz, M.; Ta, P.; Khalili, B.; Weihe, C.; Phillips, M.; Aronson, E.; Lulow, M.; Long, J.; Kimball, S. Identifying mechanisms for successful ecological restoration with salvaged topsoil in coastal sage scrub communities. Diversity 2020, 12, 150. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Lee, S.; Young, J.P.W. Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 2008, 65, 339–349. [Google Scholar] [CrossRef] [Green Version]
- Berry, D.; Mahfoudh, K.B.; Wagner, M.; Loy, A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl. Environ. Microbiol. 2011, 78, 612. [Google Scholar] [CrossRef] [Green Version]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. 2012, 27, 1–28. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst-a web-based tool for comprehensive statistical, visual, and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, 180–188. Available online: http://www.microbiomeanalyst.ca/faces/home.xhtml (accessed on 1 October 2019). [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-2. 2018. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 1 October 2019).
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R Package Version 0.1.7. 2018. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 1 October 2019).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 5–7. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 1 October 2019).
- Öpik, M.; Vanatoa, A.; Vanatoa, E.; Moora, M.; Davison, J.; Kalwij, J.M.; Reier, Ü.; Zobel, M. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytologist 2010, 188, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.N.; Singh, A.K.; Syiem, M.B. Chapter 23—Plant growth-promoting abilities in Cyanobacteria. In Cyanobacteria; Mishra, A.K., Tiwari, D.N., Rai, A.N., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 459–476. [Google Scholar]
- DeBruyn, J.; Nixon, L.; Fawaz, M.; Johnson, M.; Radosevich, M. Global Biogeography and Quantitative Season Dynamics of Gemmatimonadetes in Soil. Appl. Environ. Microbiol. 2011, 77, 295–300. [Google Scholar]
- Cha, J.Y.; Han, S.; Hong, H.J.; Cho, H.; Kim, D.; Kwon, Y.; Kwon, S.; Crusemann, M.; Lee, Y.B.; Kim, J.F.; et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016, 10, 119–129. [Google Scholar]
- Qiu, L.; Zhang, Q.; Zhu, H.; Reich, P.B.; Banerjee, S.; van der Heijden, M.G.A.; Sadowsky, M.J.; Ishii, S.; Jia, X.; Shao, M.; et al. Erosion reduces soil microbial diversity, network complexity and multifunctionality. ISME J. 2021, 15, 2474–2489. [Google Scholar] [CrossRef]
- Wang, M.; Li, S.; Chen, S.; Meng, N.; Li, X.; Zheng, H.; Zhao, C.; Wang, D. Manipulation of the rhizosphere bacterial community by biofertilizers is associated with mitigation of cadmium phytotoxicity. Sci. Total Environ. 2019, 649, 413–421. [Google Scholar] [CrossRef]
- Cao, J.; Wang, H.; Holden, N.M.; Adamowski, J.F.; Biswas, A.; Zhang, X.; Feng, Q. Soil properties and microbiome of annual and perennial cultivated grasslands on the Qinghai-Tibetan plateau. Land Degrad. Dev. 2021, 32, 5306–5321. [Google Scholar]
- Kirk, A.; Fox, S.; Entz, M.; Tenuta, M. Preliminary findings on the arbuscular mycorrhizal colonization of organic wheat. In Proceedings of the 16th IFOAM Organic World Congress, Modena, Italy, 19 June 2008. [Google Scholar]
- Weber, S.E.; Diez, J.M.; Andrews, L.V.; Goulden, M.L.; Aronson, E.L.; Allen, M.F. Responses of arbuscular mycorrhizal fungi to multiple coinciding global change drivers. Fungal Ecol. 2019, 40, 62–71. [Google Scholar] [CrossRef]
- Schussler, A. Molecular phylogeny, taxonomy, and evolution of Geoshiphon pyriformis and arbuscular mycorrhizal fungi. Plant Soil 2002, 241, 75–83. [Google Scholar] [CrossRef]
- Gong, M.; Tang, M.; Chen, H.; Zhang, Q.; Feng, X. Effects of two Glomus species on the growth and physiological performance of Sophora davidii seedlings under water stress. New For. 2013, 44, 399–408. [Google Scholar] [CrossRef]
- Berruti, A.; Lumini, E.; Balestrini, R.; Bianciotto, V. Arbuscular mycorrhizal fungi as natural biofertilizers: Let’s benefit from past successes. Front. Microbiol. 2016, 6, 1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jack, C.N.; Petipas, R.H.; Cheeke, T.E.; Rowland, J.L.; Friesen, M.L. Microbial inoculants: Silver bullet or microbial Jurassic park? Trends Microbiol. 2021, 29, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Pawar, P.B.; Khadilkar, J.P.; Kulkarni, M.V.; Melo, J.S. Compariative assessment using Glomus mosseae and NPK fertigation to evaluate its role in enhancing growth and yield of groundnut (Arachis hypogaea L.). J. Plant Nutr. 2020, 43, 1697–1711. [Google Scholar] [CrossRef]
- Van der Heijden, M.G.A.; Boller, T.; Wiemken, A.; Sanders, I.R. Different arbuscular mycorrhizal fungal species are potential determinantes of plant community structure. Ecology 1998, 79, 2082–2091. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pickett, B.; Irvine, I.C.; Arogyaswamy, K.; Maltz, M.R.; Shulman, H.; Aronson, E.L. Identifying and Remediating Soil Microbial Legacy Effects of Invasive Grasses for Restoring California Coastal Sage Scrub Ecosystems. Diversity 2022, 14, 1095. https://doi.org/10.3390/d14121095
Pickett B, Irvine IC, Arogyaswamy K, Maltz MR, Shulman H, Aronson EL. Identifying and Remediating Soil Microbial Legacy Effects of Invasive Grasses for Restoring California Coastal Sage Scrub Ecosystems. Diversity. 2022; 14(12):1095. https://doi.org/10.3390/d14121095
Chicago/Turabian StylePickett, Brooke, Irina C. Irvine, Keshav Arogyaswamy, Mia R. Maltz, Hannah Shulman, and Emma L. Aronson. 2022. "Identifying and Remediating Soil Microbial Legacy Effects of Invasive Grasses for Restoring California Coastal Sage Scrub Ecosystems" Diversity 14, no. 12: 1095. https://doi.org/10.3390/d14121095
APA StylePickett, B., Irvine, I. C., Arogyaswamy, K., Maltz, M. R., Shulman, H., & Aronson, E. L. (2022). Identifying and Remediating Soil Microbial Legacy Effects of Invasive Grasses for Restoring California Coastal Sage Scrub Ecosystems. Diversity, 14(12), 1095. https://doi.org/10.3390/d14121095