Paeonol Ameliorates Ulcerative Colitis in Mice by Modulating the Gut Microbiota and Metabolites
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animal Experiments and Sample Collection
2.3. Disease Activity Index (DAI)
2.4. Histological Examination
2.5. Transmission Electron Microscopy (TEM)
2.6. ELISA Analysis
2.7. Immunohistochemistry
2.8. RT-qPCR
2.9. Western Blotting
2.10. Gut Microbiota Profiling by 16S rRNA Sequencing
2.11. Targeted Fecal Metabolomics Analysis
2.12. Statistical Analysis
3. Results
3.1. Pae Attenuates the Effect of DSS-Induced UC in Mice
3.2. Pae Restores Intestinal Barrier Function in DSS-Induced UC
3.3. Pae Attenuates DSS-Induced Dysregulation of the Gut Microbiota in Mice with UC
3.4. Effect of Pae on Fecal Metabolic Disorders Caused by DSS-Induced UC in Mice
3.5. Pae Improves UC via Pathways Involving Gut Microbiota-BAs-FXR/FGF15 Signaling
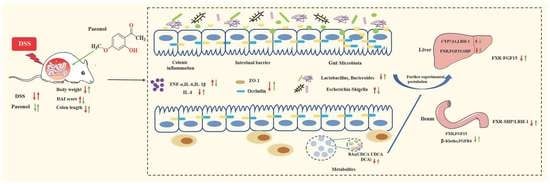
3.6. Integrated Map of the Mechanism of Pae in Ameliorating UC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sartor, R.B. Mechanisms of disease: Pathogenesis of crohn’s disease and ulcerative colitis. Nat. Clin. Pract. Gastr. 2006, 3, 390–407. [Google Scholar] [CrossRef] [PubMed]
- Panwala, C.M.; Jones, J.C.; Viney, J.L. A novel model of inflammatory bowel disease: Mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J. Immunol. 1998, 161, 5733–5744. [Google Scholar] [PubMed]
- Li, C.; Dong, N.; Wu, B.; Mo, Z.; Xie, J.; Lu, Q. Dihydroberberine, an isoquinoline alkaloid, exhibits protective effect against dextran sulfate sodium-induced ulcerative colitis in mice. Phytomedicine 2021, 90, 153631. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Mytton, J.; Evison, F.; Kamarajah, S.K.; Reece, J.; Iqbal, T.; Cooney, R.; Thompson, F.; Walmsley, M.; Ferguson, J.; et al. A nationwide population-based evaluation of mortality and cancer-risk in patients with ulcerative colitis/primary sclerosing cholangitis—young age at diagnosis and the unmet need to reduce mortality. J. Hepatol. 2018, 68, S220–S221. [Google Scholar] [CrossRef]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Su, L.; Ma, F.; An, Z.; Ji, X.; Zhang, P.; Yue, Q.; Zhao, C.; Sun, X.; Li, K.; Li, B.; et al. The metabolites of lactobacillus fermentum f-b9-1 relieved dextran sulfate sodium-induced experimental ulcerative colitis in mice. Front. Microbiol. 2022, 13, 865925. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.; Yu, Z.; Zhang, L.; Duo, T.; Zhao, Y.; Qin, W.; Yang, W.; Ma, L. Berberine ameliorates dextran sulfate sodium-induced ulcerative colitis and inhibits the secretion of gut lysozyme via promoting autophagy. Metabolites 2022, 12, 676. [Google Scholar] [CrossRef] [PubMed]
- Citi, S. Intestinal barriers protect against disease. Science 2018, 359, 1097–1098. [Google Scholar] [CrossRef] [PubMed]
- Mehandru, S.; Colombel, J.-F. The intestinal barrier, an arbitrator turned provocateur in ibd. Nat. Rev. Gastro. Hepat. 2021, 18, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Kiernan, M.G.; Coffey, J.C.; Sahebally, S.M.; Tibbitts, P.; Lyons, E.M.; O'Leary, E.; Owolabi, F.; Dunne, C.P. Systemic molecular mediators of inflammation differentiate between crohn’s disease and ulcerative colitis, implicating threshold levels of il-10 and relative ratios of pro-inflammatory cytokines in therapy. J. Crohns. Colitis. 2020, 14, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.-A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Imhann, F.; Vila, A.V.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Haesler, R.; et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology 2017, 153, 1504–1516.e2. [Google Scholar] [CrossRef] [Green Version]
- Gnewuch, C.; Liebisch, G.; Langmann, T.; Dieplinger, B.; Mueller, T.; Haltmayer, M.; Dieplinger, H.; Zahn, A.; Stremmel, W.; Rogler, G.; et al. Serum bile acid profiling reflects enterohepatic detoxification state and intestinal barrier function in inflammatory bowel disease. World J. Gastroentero. 2009, 15, 3134–3141. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, gpcr, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.L.; Takeda, K.; Sundrud, M.S. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal. Immunol. 2019, 12, 851–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modica, S.; Petruzzelli, M.; Bellafante, E.; Murzilli, S.; Salvatore, L.; Celli, N.; Di Tullio, G.; Palasciano, G.; Moustafa, T.; Halilbasic, E.; et al. Selective activation of nuclear bile acid receptor fxr in the intestine protects mice against cholestasis. Gastroenterology 2012, 142, 355–365.e351–354. [Google Scholar] [CrossRef] [PubMed]
- Arrese, M.; Karpen, S.J. New horizons in the regulation of bile acid and lipid homeostasis: Critical role of the nuclear receptor fxr as an intracellular bile acid sensor. Gut 2001, 49, 465–466. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, T.; Choi, M.; Moschetta, A.; Peng, L.; Cummins, C.L.; McDonald, J.G.; Luo, G.; Jones, S.A.; Goodwin, B.; Richardson, J.A.; et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005, 2, 217–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsubara, T.; Li, F.; Gonzalez, F.J. Fxr signaling in the enterohepatic system. Mol. Cell Endocrinol. 2013, 368, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Cao, L.; Jiang, C.; Xie, Y.; Cheng, X.; Krausz, K.W.; Qi, Y.; Sun, L.; Shah, Y.M.; Gonzalez, F.J.; et al. Pparα-ugt axis activation represses intestinal fxr-fgf15 feedback signalling and exacerbates experimental colitis. Nat. Commun. 2014, 5, 4573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.-H.; Liu, F.; Zhu, X.-R.; Suo, F.-Y.; Jia, Z.-j.; Yao, S.-K. Altered profiles of fecal bile acids correlate with gut microbiota and inflammatory responses in patients with ulcerative colitis. World J. Gastroentero. 2021, 27, 3609–3629. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.-L.; Jia, Y.-Q.; Zhang, X.-S.; Yuan, Z.-W.; Ji, P.; Hu, J.-J.; Wei, Y.-M. Baitouweng tang ameliorates dss-induced ulcerative colitis through the regulation of the gut microbiota and bile acids via pathways involving fxr and tgr5. Biomed. Pharmacother. 2021, 137, 111320. [Google Scholar] [CrossRef] [PubMed]
- Machado, A.P.d.F.; Geraldi, M.V.; do Nascimento, R.d.P.; Moya, A.M.T.M.; Vezza, T.; Diez-Echave, P.; Juan Galvaz, J.; Cazarin, C.B.B.; Marostica Junior, M.R. Polyphenols from food by-products: An alternative or complementary therapy to IBD conventional treatments. Food Res. Int. 2021, 140, 110018. [Google Scholar] [CrossRef] [PubMed]
- Chaouch, M.A.; Benvenuti, S. The role of fruit by-products as bioactive compounds for intestinal health. Foods 2020, 9, 1716. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, D.C.; Liu, L.F. Paeonol: Pharmacological effects and mechanisms of action. Int. Immunopharmacol. 2019, 72, 413–421. [Google Scholar] [CrossRef]
- Adki, K.M.; Murugesan, S.; Kulkarni, Y.A. In silico and in vivo toxicological evaluation of paeonol. Chem. Biodivers. 2020, 17, e2000422. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhu, R.D.; Cao, G.M.; Du, J.C.; Liu, X.; Diao, L.Z.; Zhang, Z.Y.; Hu, Y.S.; Liu, X.H.; Shi, J.B. Discovery of novel paeonol-based derivatives against skin inflammation in vitro and in vivo. J. Enzym. Inhib. Med. Ch. 2022, 37, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Anti-inflammatory and analgesic effects of paeonol in carrageenan-evoked thermal hyperalgesia. Brit. J. Pharmacol. 2003, 139, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.-Y.; Tsai, T.-H.; Ho, T.-Y.; Lin, Y.-W.; Cheng, C.-Y.; Hsieh, C.-L. Neuroprotective effect of paeonol mediates anti-inflammation via suppressing toll-like receptor 2 and toll-like receptor 4 signaling pathways in cerebral ischemia-reperfusion injured rats. Evid.-Based Compl. Alt. 2016, 2016, 3704647. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Qin, Y.; Chen, C.; Guo, X. Beneficial effects exerted by paeonol in the management of atherosclerosis. Oxid. Med. Cell. Longev. 2018, 2018, 1098617. [Google Scholar]
- Zong, S.-y.; Pu, Y.-q.; Xu, B.-l.; Zhang, T.; Wang, B. Study on the physicochemical properties and anti-inflammatory effects of paeonol in rats with tnbs-induced ulcerative colitis. Int. Immunopharmacol. 2017, 42, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Pan, M.; Zhang, C.; Wang, C.; Ma, K.; Yan, G.; Wang, T.; Wu, D.; Shao, J. Paeonol alleviates dextran sodium sulfate induced colitis involving candida albicans-associated dysbiosis. Med. Mycol. 2021, 59, 335–344. [Google Scholar] [CrossRef]
- He, X.; Liu, J.; Long, G.; Xia, X.H.; Liu, M. 2,3,5,4'-tetrahydroxystilbene-2-o-β-d-glucoside, a major bioactive component from polygoni multiflori radix (heshouwu) suppresses dss induced acute colitis in balb/c mice by modulating gut microbiota. Biomed. Pharmacother. 2021, 137, 111420. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Z.; Mai, P.; Hao, Y.; Wang, Z.; Wang, J. Quinoa bran soluble dietary fiber ameliorates dextran sodium sulfate induced ulcerative colitis in balb/c mice by maintaining intestinal barrier function and modulating gut microbiota. Int. J. Biol. Macromol. 2022, 216, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Yuan, Z.W.; Qu, C.; Yu, X.T.; Huang, T.; Chen, P.V.; Su, Z.R.; Dou, Y.X.; Wu, J.Z.; Zeng, H.F.; et al. Palmatine ameliorated murine colitis by suppressing tryptophan metabolism and regulating gut microbiota. Pharmacol. Res. 2018, 137, 34–46. [Google Scholar] [CrossRef]
- Bang, B.; Lichtenberger, L.M. Methods of inducing inflammatory bowel disease in mice. Curr. Protoc. Pharmacol. 2016, 72, 5.58.51–55.58.42. [Google Scholar] [CrossRef]
- Jing, W.; Safarpour, Y.; Zhang, T.; Guo, P.; Chen, G.; Wu, X.; Fu, Q.; Wang, Y. Berberine upregulates p-glycoprotein in human caco-2 cells and in an experimental model of colitis in the rat via activation of nrf2-dependent mechanisms. J. Pharmacol. Exp. Ther. 2018, 366, 332–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.; Leong, R.W.; Wasinger, V.C.; Ip, M.; Yang, M.; Tri Giang, P. Impaired intestinal permeability contributes to ongoing bowel symptoms in patients with inflammatory bowel disease and mucosal healing. Gastroenterology 2017, 153, 723.e721–731.e721. [Google Scholar] [CrossRef]
- Feng, W.; Liu, J.; Tan, Y.; Ao, H.; Wang, J.; Peng, C. Polysaccharides from atractylodes macrocephala koidz. Ameliorate ulcerative colitis via extensive modification of gut microbiota and host metabolism. Food Res. Int. 2020, 138, 109777. [Google Scholar] [CrossRef]
- Chen, X.-q.; Lv, X.-y.; Liu, S.-j. Baitouweng decoction alleviates dextran sulfate sodium-induced ulcerative colitis by regulating intestinal microbiota and the il-6/stat3 signaling pathway. J. Ethnopharmacol. 2021, 265, 113357. [Google Scholar]
- Bromke, M.A.; Krzystek-Korpacka, M. Bile acid signaling in inflammatory bowel disease. Int. J. Mol. Sci. 2021, 22, 9096. [Google Scholar] [CrossRef]
- Lavelle, A.; Nancey, S.; Reimund, J.-M.; Laharie, D.; Marteau, P.; Treton, X.; Allez, M.; Roblin, X.; Malamut, G.; Oeuvray, C.; et al. Fecal microbiota and bile acids in ibd patients undergoing screening for colorectal cancer. Gut Microbes 2022, 14, 2078620. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [PubMed] [Green Version]
- Qu, L.; Liu, C.; Ke, C.; Zhan, X.; Li, L.; Xu, H.; Xu, K.; Liu, Y. Atractylodes lancea rhizoma attenuates dss-induced colitis by regulating intestinal flora and metabolites. Am. J. Chin. Med. 2022, 50, 525–552. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, S.; Cao, H.; Shen, P.; Liu, J.; Fu, Y.; Cao, Y.; Zhang, N. The protective role of phloretin against dextran sulfate sodium-induced ulcerative colitis in mice. Food Funct. 2019, 10, 422–431. [Google Scholar] [CrossRef]
- Lechuga, S.; Ivanov, A.I. Disruption of the epithelial barrier during intestinal inflammation: Quest for new molecules and mechanisms. BBA-Mol. Cell. Res. 2017, 1864, 1183–1194. [Google Scholar] [CrossRef]
- Suzuki, T. Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 2013, 70, 631–659. [Google Scholar] [PubMed]
- Fischer, A.; Gluth, M.; Pape, U.; Wiedenmann, B.; Theuring, F.; Baumgart, D. Adalimumab prevents barrier dysfunction and antagonizes distinct effects of tnf-α on tight junction proteins and signaling pathways in intestinal epithelial cells. Am. J. Physiol.-Gastr. L. 2013, 304, G970–G979. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Jiang, W.; Jiang, J.; Zhao, J.; Liu, Y.; Zhang, Y.; Zhou, X.; Feng, L. Dietary choline deficiency and excess induced intestinal inflammation and alteration of intestinal tight junction protein transcription potentially by modulating nf-κb, stat and p38 mapk signaling molecules in juvenile jian carp. Fish Shellfish. Immun. 2016, 58, 462–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Yoshinaga, N.; Tanabe, S. Interleukin-6 (il-6) regulates claudin-2 expression and tight junction permeability in intestinal epithelium. J. Biol. Chem. 2011, 286, 31263–31271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinugasa, T.; Sakaguchi, T.; Gu, X.B.; Reinecker, H.C. Claudins regulate the intestinal barrier in response to immune mediators. Gastroenterology 2000, 118, 1001–1011. [Google Scholar] [CrossRef]
- Liao, Z.; Xie, Y.; Zhou, B.; Zou, B.; Xiao, D.; Liu, W.; Cai, Y.; Liu, D.; Liao, Q.; Xie, Z. Berberine ameliorates colonic damage accompanied with the modulation of dysfunctional bacteria and functions in ulcerative colitis rats. Appl. Microbiol. Biot. 2020, 104, 1737–1749. [Google Scholar] [CrossRef]
- Zhao, H.; Cheng, N.; Zhou, W.; Chen, S.; Wang, Q.; Gao, H.; Xue, X.; Wu, L.; Cao, W. Honey polyphenols ameliorate dss-induced ulcerative colitis via modulating gut microbiota in rats. Mol. Nutr. Food Res. 2019, 63, e1900638. [Google Scholar] [CrossRef]
- Dong, S.; Zhu, M.; Wang, K.; Zhao, X.; Hu, L.; Jing, W.; Lu, H.; Wang, S. Dihydromyricetin improves dss-induced colitis in mice via modulation of fecal-bacteria-related bile acid metabolism. Pharmacol. Rep. 2021, 171, 105767. [Google Scholar] [CrossRef]
- Serena, C.; Queipo-Ortuno, M.; Millan, M.; Sanchez-Alcoholado, L.; Caro, A.; Espina, B.; Menacho, M.; Bautista, M.; Monfort-Ferre, D.; Terron-Puig, M.; et al. Microbial signature in adipose tissue of crohn's disease patients. J. Clin. Med. 2020, 9, 2448. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, S.; Zhang, X.; Cao, X.; Liu, D.; Zhu, Y.; Ye, S.; Xu, Z.; Liao, Q.; Hong, Y.; et al. Integrated microbiome-metabolomics analysis reveals the potential therapeutic mechanism of zuo-jin-wan in ulcerative colitis. Phytomedicine 2022, 98, 153914. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; Leleiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Deng, J.; Chen, X.; Wan, S.S.; Wang, Y.; Cao, Q. High incidence and morbidity of clostridium difficile infection among hospitalized patients with inflammatory bowel disease: A prospective observational cohort study. J. Digest. Dis. 2019, 20, 460–466. [Google Scholar] [CrossRef]
- Peng, L.; Gao, X.; Nie, L.; Xie, J.; Dai, T.; Shi, C.; Tao, L.; Wang, Y.; Tian, Y.; Sheng, J. Astragalin attenuates dextran sulfate sodium (dss)-induced acute experimental colitis by alleviating gut microbiota dysbiosis and inhibiting nf-κb activation in mice. Front. Immunol. 2020, 11, 2058. [Google Scholar] [CrossRef]
- Pang, B.; Jin, H.; Liao, N.; Li, J.; Jiang, C.; Shi, J. Vitamin a supplementation ameliorates ulcerative colitis in gut microbiota-dependent manner. Food Res. Int. 2021, 148, 110568. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic t-reg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastro. Hepat. 2018, 15, 111–128. [Google Scholar]
- Liu, F.; Wang, X.; Li, D.; Cui, Y.; Li, X. Apple polyphenols extract alleviated dextran sulfate sodium-induced ulcerative colitis in c57bl/6 male mice by restoring bile acid metabolism disorder and gut microbiota dysbiosis. Phytother. Res. 2021, 35, 1468–1485. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, F.; Yuan, W.; Wei, Y.; Wei, M.; Zhou, Y.; Yang, Y.; Chang, Y.; Wu, X. Hepatic cholesterol accumulation ascribed to the activation of ileum fxr-fgf15 pathway inhibiting hepatic cyp7a1 in high-fat diet-induced obesity rats. Life Sci. 2019, 232, 116638. [Google Scholar] [CrossRef] [PubMed]
- Tayyar, A.T.; Tayyar, A.; Kozali, S.; Karakus, R.; Koroglu, N.; Yuksel, I.T.; Yildirim, G.Y.; Dag, I.; Eroglu, M. Evaluation of fgf-19 and beta-klotho as biomarkers in patients with intrahepatic cholestasis of pregnancy. Arch. Med. Sci. 2019, 15, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Goyal, N.; Rana, A.; Bijjem, K.R.V.; Kumar, P. Effect of chenodeoxycholic acid and sodium hydrogen sulfide in dinitro benzene sulfonic acid (dnbs)—induced ulcerative colitis in rats. Pharmacol. Rep. 2015, 67, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Lajczak-McGinley, N.K.; Porru, E.; Fallon, C.M.; Smyth, J.; Curley, C.; McCarron, P.A.; Tambuwala, M.M.; Roda, A.; Keely, S.J. The secondary bile acids, ursodeoxycholic acid and lithocholic acid, protect against intestinal inflammation by inhibition of epithelial apoptosis. Physiol. Rep. 2020, 8, e14456. [Google Scholar] [CrossRef] [PubMed]
- Weylandt, K.; Kang, J.; Wiedenmann, B.; Baumgart, D. Lipoxins and resolvins in inflammatory bowel disease. Inflamm. Bowel Dis. 2007, 13, 797–799. [Google Scholar] [CrossRef]
- Strauch, E.; Yamaguchi, J.; Bass, B.; Wang, J. Bile salts regulate intestinal epithelial cell migration by nuclear factor-kappa b-induced expression of transforming growth factor-beta. J. Am. Coll. Surg. 2003, 197, 974–984. [Google Scholar] [CrossRef]







| Body Weight Loss | Stool Consistency | Presence of Gross Bleeding or Bloodstain | Score |
|---|---|---|---|
| No loss | Normal | Negative | 0 |
| 1–5% | 1 | ||
| 5–10% | Loose stools | Positive | 2 |
| 10–15% | 3 | ||
| Over 15% | Diarrhea | Gross rectal bleeding | 4 |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| FGF15 | ACCAGAAACCCTCAAACT | CTACATCCTCCACCATCC |
| FXR | CCATTTACAGGCTACGGA | ACTTGAGGAAACGGGACA |
| SHP | CCCAGCAAGGACACTGAGCAAG | CCTCGAAGGTCACAGCAT |
| LRH-1 | CTGAGTCAATGATGGGTTA | CTTTTCTTGCCTGTTTCG |
| β-KLOTHO | ACCATTTGCTCATTTCTCG | ACTCTGCTGTGGCCTTTC |
| FGFR4 | TGGGCTAATGAGGGAGTG | AGGCGGAGGTCAAGGTAC |
| GAPDH | CCCAGCAAGGACACTGAGCAAG | GGTCTGGGATGGAAATTGTGAGGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Li, H.; Zhang, P.; Yue, S.; Zhai, B.; Zou, J.; Cheng, J.; Zhao, C.; Guo, D.; Wang, J. Paeonol Ameliorates Ulcerative Colitis in Mice by Modulating the Gut Microbiota and Metabolites. Metabolites 2022, 12, 956. https://doi.org/10.3390/metabo12100956
Zheng J, Li H, Zhang P, Yue S, Zhai B, Zou J, Cheng J, Zhao C, Guo D, Wang J. Paeonol Ameliorates Ulcerative Colitis in Mice by Modulating the Gut Microbiota and Metabolites. Metabolites. 2022; 12(10):956. https://doi.org/10.3390/metabo12100956
Chicago/Turabian StyleZheng, Jiahui, Huan Li, Pei Zhang, Shijun Yue, Bingtao Zhai, Junbo Zou, Jiangxue Cheng, Chongbo Zhao, Dongyan Guo, and Jing Wang. 2022. "Paeonol Ameliorates Ulcerative Colitis in Mice by Modulating the Gut Microbiota and Metabolites" Metabolites 12, no. 10: 956. https://doi.org/10.3390/metabo12100956
APA StyleZheng, J., Li, H., Zhang, P., Yue, S., Zhai, B., Zou, J., Cheng, J., Zhao, C., Guo, D., & Wang, J. (2022). Paeonol Ameliorates Ulcerative Colitis in Mice by Modulating the Gut Microbiota and Metabolites. Metabolites, 12(10), 956. https://doi.org/10.3390/metabo12100956