Improvement of Cucurbitacin B Content in Cucumis melo Pedicel Extracts by Biotransformation Using Recombinant β-Glucosidase
Abstract
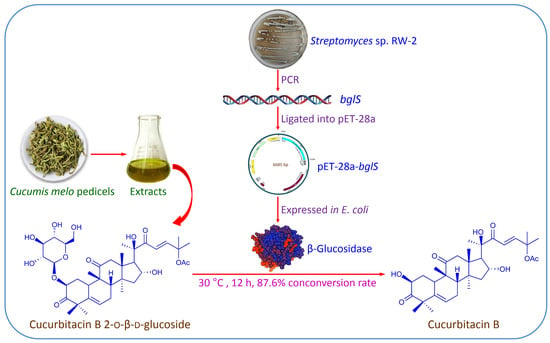
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Enzymes
2.2. Strains and Plasmids
2.3. Construction of Recombinant E. coli
2.4. Expression of Recombinant β-Glucosidase
2.5. Enzyme Essay
2.6. Biotransformation Process
2.7. Analysis of Biotransformation Products
3. Results and Discussion
3.1. Cloning of bglS and Sequence Analysis
3.2. Expression of bglS in E. coli
3.3. Biotranformation of CuBg by Recombinant β-glucosidase
3.4. Biotransformation of C. melo Pedicel Extracts by Recombinant β-Glucosidase
4. Conclusions
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, J.C.; Chiu, M.H.; Nie, R.L.; Cordell, G.A.; Qiu, S.X. Cucurbitacins and cucurbitane glycosides: Structures and biological activities. Nat. Prod. Rep. 2005, 22, 386–399. [Google Scholar] [CrossRef] [PubMed]
- Miró, M. Cucurbitacins and their pharmacological effects. Phytother. Res. 1995, 9, 159–168. [Google Scholar] [CrossRef]
- Aeri, V.; Kaushik, U.; Mir, S. Cucurbitacins—An insight into medicinal leads from nature. Pharmacogn. Rev. 2015, 9, 12–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Fang, X.; He, C.; Li, P.; Xiao, F.; Wang, Y.; Chen, M. Cucurbitacins: A systematic review of the phytochemistry and anticancer activity. Am. J. Chin. Med. 2015, 43, 1331–1350. [Google Scholar] [CrossRef] [PubMed]
- Garg, S.; Kaul, S.; Wadhwa, R. Cucurbitacin B and cancer intervention: Chemistry, biology and mechanisms (Review). Int. J. Oncol. 2017, 52, 19–37. [Google Scholar] [CrossRef]
- Hussain, H.; Green, I.R.; Saleem, M.; Khattak, K.F.; Irshad, M.; Ali, M. Cucurbitacins as anticancer agents: A patent review. Recent Pat. Anti-Cancer Drug Discov. 2019, 14, 133–143. [Google Scholar] [CrossRef]
- Piao, X.-M.; Gao, F.; Zhu, J.-X.; Wang, L.-J.; Zhao, X.; Li, X.; Sheng, M.M.; Zhang, Y. Cucurbitacin B inhibits tumor angiogenesis by triggering the mitochondrial signaling pathway in endothelial cells. Int. J. Mol. Med. 2018, 42, 1018–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, W.G.; Tang, L.; Shan, H.F.; Zhan, Z.J. A Method for Extraction and Separation of Cucurbitacin B. CN101759746A, 30 June 2010. [Google Scholar]
- Su, L.H. A Method for Extraction of Cucurbitacin B. CN101974055A, 16 February 2011. [Google Scholar]
- Chao, P.-D.L.; Hsiu, S.-L.; Hou, Y.-C. Bioavailability, metabolism, and pharmacokinetics of glycosides in Chinese herbs. ACS Symp. Ser. 2006, 925, 212–223. [Google Scholar] [CrossRef]
- Xu, X.; Tang, L.; Shan, H.F.; Wang, Z.Q.; Shan, W.G. Study on extraction of cucurbitacin B from the pedicel of Cucumis melo L. by acid hydrolysis. Adv. Mater. Res. 2013, 704, 61–65. [Google Scholar] [CrossRef]
- Mei, J.; Li, S.; Jin, H.; Tang, L.; Yi, Y.; Wang, H.; Ying, G. A biotransformation process for the production of cucurbitacin B from its glycoside using a selected Streptomyces sp. Bioprocess. Biosyst. Eng. 2016, 39, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, C.-C.; Zhang, H.; Zhang, L.-X.; Liu, Z.; Sun, G.-Z.; Lei, J.; Qin, Y.-X.; Zheng, Y.-N.; Li, X.; Pan, H.-Y. Biotransformation of ginsenoside Rf to Rh1 by recombinant β-glucosidase. Molecules 2009, 14, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Cardillo, A.B.; Perassolo, M.; Sartuqui, M.; Talou, J.R.; Giulietti, A.M. Production of tropane alkaloids by biotransformation using recombinant Escherichia coli whole cells. Biochem. Eng. J. 2017, 125, 180–189. [Google Scholar] [CrossRef]
- Park, H.L.; Lee, J.C.; Lee, K.; Lee, J.M.; Nam, H.J.; Bhoo, S.H.; Lee, T.H.; Lee, S.-W.; Cho, M.-H. Biochemical characterization of a flavonoid o-methyltransferase from Perilla leaves and its application in 7-methoxyflavonoid production. Molecules 2020, 25, 4455. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.Z.; Medjebouri, S.; Liu, Q.; Park, H.Y.; Kim, G.-R.; Im, W.-T. Efficient production of various minor ginsenosides from PPD- and PPT-type major ginsenosides using a single recombinant BglFc isolated from Flavobacterium chilense. Biotechnol. Bioprocess. Eng. 2021, 26, 232–246. [Google Scholar] [CrossRef]
- Koko, M.; Sami, R.; Muhoza, B.; Khojah, E.; Mansour, A. Promising pathway of thermostable mannitol dehydrogenase (MtDH) from Caldicellulosiruptor hydrothermalis 108 for d-mannitol synthesis. Separations 2021, 8, 76. [Google Scholar] [CrossRef]
- Brunelle, J.L.; Green, R. One-dimensional SDS-polyacrylamide gel electrophoresis (1D SDS-PAGE). Methods Enzymol. 2014, 541, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Cruz Rodríguez, A.; Sánchez Esperanza, F.A.; Pérez-Campos, E.; Hernández-Huerta, M.T.; Pérez-Campos Mayoral, L.; Matias-Cervantes, C.A.; Martínez Barras, A.; Mayoral-Andrade, G.; Santos Pineda, L.Á.; Díaz Barrita, A.J.; et al. Aggregation and molecular properties of β-glucosidase isoform II in chayote (Sechium edule). Molecules 2020, 25, 1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, J.; Wu, X.; Zheng, S.; Chen, X.; Huang, Z.; Wu, Y. Improvement of Cucurbitacin B Content in Cucumis melo Pedicel Extracts by Biotransformation Using Recombinant β-Glucosidase. Separations 2021, 8, 138. https://doi.org/10.3390/separations8090138
Mei J, Wu X, Zheng S, Chen X, Huang Z, Wu Y. Improvement of Cucurbitacin B Content in Cucumis melo Pedicel Extracts by Biotransformation Using Recombinant β-Glucosidase. Separations. 2021; 8(9):138. https://doi.org/10.3390/separations8090138
Chicago/Turabian StyleMei, Jianfeng, Xia Wu, Sujing Zheng, Xiang Chen, Zhuliang Huang, and Yichun Wu. 2021. "Improvement of Cucurbitacin B Content in Cucumis melo Pedicel Extracts by Biotransformation Using Recombinant β-Glucosidase" Separations 8, no. 9: 138. https://doi.org/10.3390/separations8090138
APA StyleMei, J., Wu, X., Zheng, S., Chen, X., Huang, Z., & Wu, Y. (2021). Improvement of Cucurbitacin B Content in Cucumis melo Pedicel Extracts by Biotransformation Using Recombinant β-Glucosidase. Separations, 8(9), 138. https://doi.org/10.3390/separations8090138